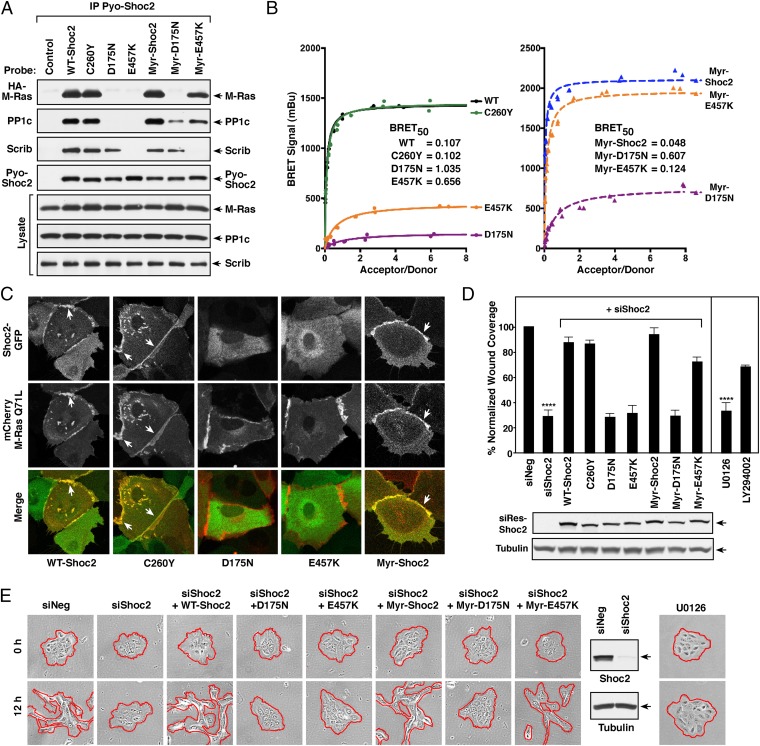
Fig. 3.
Analysis of Shoc2 mutant proteins: protein interactions, localization, and migratory properties. (A) Pyo-Shoc2 proteins were immunoprecipitated from lysates of MCF10A cells stably expressing HA-M-RasQ71L together with the indicated Pyo-Shoc2 proteins. Pyo-Shoc2 complexes were probed for the presence of HA-M-Ras, PP1c, Scribble (Scrib), and Pyo-Shoc2. (B) BRET analysis of binding interactions between the indicated Shoc2-Rluc8 proteins and Venus-M-RasQ71L. (C) Live cell imaging of cells expressing the indicated GFP-Shoc2 proteins and mCherry-M-RasQ71L. (D and E) Control MCF10A cells or lines stably expressing siShoc2-resistant GFP-Shoc2 proteins were transfected with control or Shoc2 siRNAs before analysis in wound-healing (D) and cell-scattering assays (E). GFP-Shoc2 and tubulin levels are also shown. Error bars represent mean ± SD, ****P > 0.0001. Red lines indicate free cell edges.