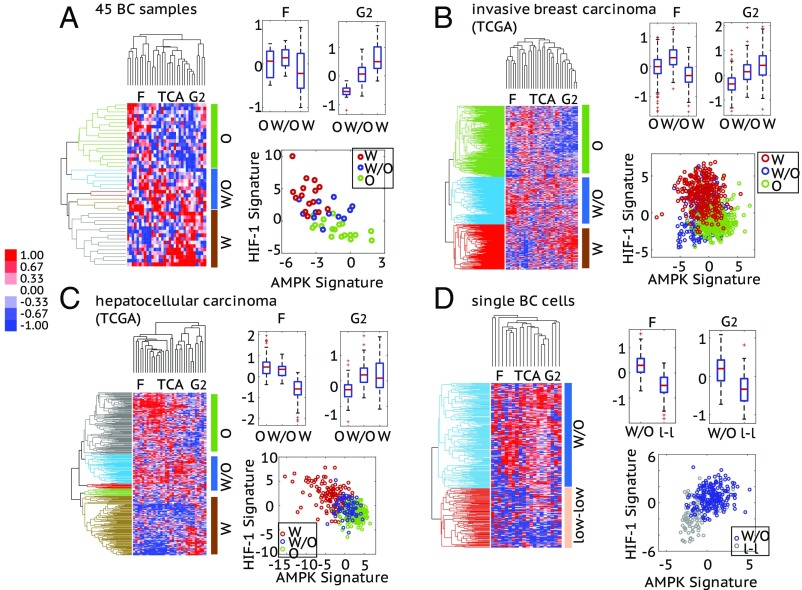
Fig. 6.
Association of the AMPK/HIF-1 activity with the metabolic pathway activity in 45 BC samples (A), 1,100 invasive breast carcinoma samples from TCGA (B), 373 HCC samples from TCGA (C), and 317 single BC cells (D). (A–C, Left) Average linkage HCA of enzyme gene expression (microarray data for A and RNA-seq data for B and C) of patient samples. The similarity metric used here is based on Pearson correlation. Each row represents a patient sample and each column represents the expression of one enzyme gene. Three major enzyme gene states, O, W/O, and W, are identified and highlighted by different colors in the dendrogram. The cutoff values to get these clusters are 0.04 for A, 0.1 for B, and 0.06 for C. (Right, Top) Box plots for the FAO (F) and glycolysis (G2) scores of the clustered patient samples. For 45 BC samples: FAO score, , , ; glycolysis score, , , . For invasive breast carcinoma: FAO score, , , ; glycolysis score, , , . For HCC: FAO score, , , ; glycolysis score, , , . (Right, Bottom) The AMPK and HIF-1 signatures of the clustered patient samples. Here, each hollow dot represents one patient sample. (D, Left) Average linkage HCA of the RNA-seq data of enzyme genes of single BC cells. The enzyme genes whose expression was detected in more than 150 single cells (approximately half of the total) were used to do clustering. The similarity metric used here is based on Pearson correlation. Each row represents a single cell and each column represents the expression of one enzyme gene. Two major enzyme gene states—W/O and low-low—are identified and highlighted by different colors in the dendrogram. The cutoff value to get these two clusters is 0.009. (Right, Top) Box plots for the F and G2 scores of cells in the W/O and low-low clusters. FAO score, ; glycolysis score, . (Right, Bottom) The AMPK and HIF-1 signatures of the single cells in the W/O cluster and the low-low cluster. For the heat maps in A–D, the names of the columns are listed as the names of the metabolism pathways and the full list of enzyme genes can be found in SI Appendix, Table S8. The TCA score of each case is shown in SI Appendix, Fig. S8. For all box plots here, P values for a balanced one-way ANOVA are calculated.