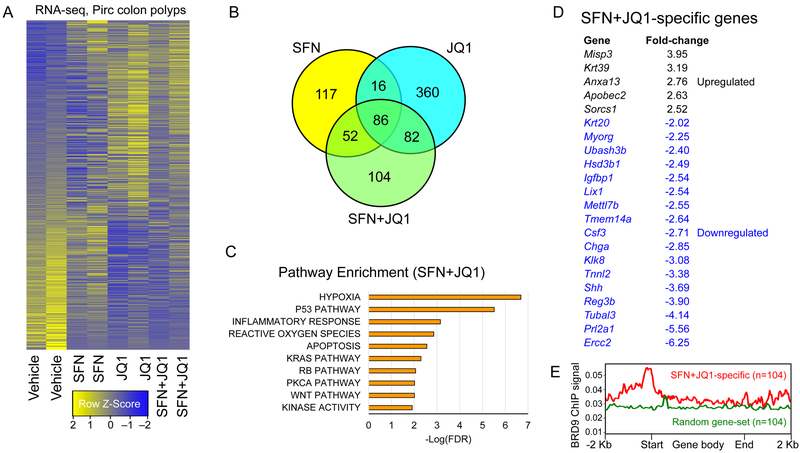
Figure 6.
RNA-seq prioritizes HDAC+BET ‘cooperativity’ genes in Pirc colon tumors. A, Heatmaps for groups analyzed in duplicate (1436 DEGs, no cutoff applied). B, Number of DEGs compared to vehicle controls. C, Pathway enrichment analysis for SFN+JQ1 DEGs. D, Highly upregulated and downregulated SFN+JQ1-specific genes. E, Human genome reference GRCh38 was interrogated using BRD9 ChIP-seq data downloaded from GSM2092891. The profile was plotted using Mmint, with the red line representing the average BRD9 signal for SFN+JQ1 ‘synergy/cooperativity’ genes. The corresponding BRD9 signal also was examined for a set of 104 randomly selected genes, not among the ‘synergy/cooperativity’ candidates (green line). Start, transcription start sites; End, transcription stop sites (n=104).