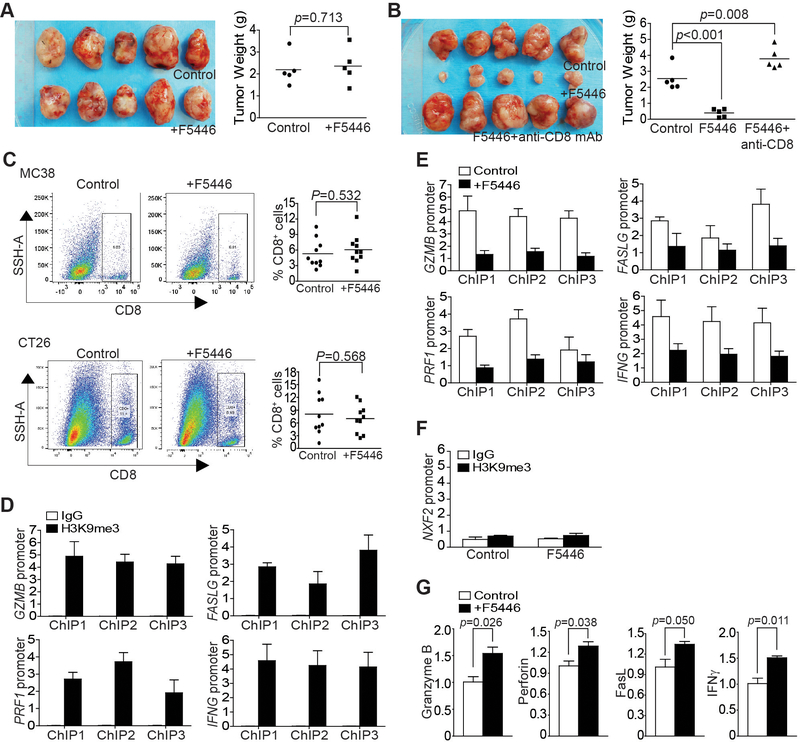
Figure 7. The SUV39H1-H3K9me3 pathway represses T-cell effector gene expression in tumor-infiltrating CTLs in vivo.
A. MC38 cells were injected to twelve Rag1 KO mice subcutaneously. Ten mice with similar sized tumors were randomly assigned into two groups at day 8, and treated with vehicle (control, n=5) or F5446 (10 mg/kg, n=5) every two days for 14 days. Left panel shows tumor images. Tumors were excised from sacrificed mice 22 days after tumor cell injection, weighed and presented at the right panel. Each dot represents tumor weight of tumor from one mouse. Differences in tumor weights between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. B. CT26 cells (2×105 cells/mouse) were injected to twenty BALB/c mice subcutaneously. Fifteen mice with similar sized tumors were randomly assigned into three groups at day 10 after tumor cell injection and treated with vehicle (control, n=5), F5446 (10 mg/kg, n=5), and F5446+anti-CD8 (n=5) every two days for 14 days. Left panel shows tumor images. Tumors were excised from sacrificed mice 24 days after tumor cell injection, weighed, and presented at the right panel. Each dot represents tumor weight of tumor from one mouse. Differences in tumor weights between control and the treatment group were determined by two-tail t test with p<0.05 as being statistically significant. C. Tumor tissues were collected from the control (two independent experiments, n=5 each) and F5446-treated (two independent experiments, n=5 each) MC38 and CT26 tumor-bearing mice, respectively, digested with a collagenase solution and stained with Zombie Violet, anti-CD45 and anti-CD8. CD45+ cells were gated from live cells (Zombie Violet- cells) and were further gated for CD8+ cells. Shown are representative dot plots (left panel). The % CD8+ T cells was quantified and pooled from the two independent experiments and presented in the right panel. Each dot represents data from one mouse. Bar: mean. Differences in % CD8+ cells between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. D. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice as shown in Fig. 6E and analyzed by ChIP with anti-H3K9me3 and gene-specific PCR primers in triplicates. ChIP 1–3 represents the three regions of the respective gene promoter amplified by ChIP PCR as shown in Fig. 3B. Column: mean; bar: SEM. E. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice from the control and F5446 treatment groups as shown in Fig. 6E. The isolated CTLs were then analyzed by ChIP with anti-H3K9me3 and gene-specific PCR primers in triplicates as indicated in Fig. 3B. The H3K9me3 was normalized to input. ChIP 1–3 indicates the ChIP PCR amplified three regions of the respective gene promoter as in Fig. 3B. Column: mean; Bar: SEM. F. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice from the control and F5446 treatment groups as shown in Fig. 6E. The isolated CTLs were then analyzed by ChIP with IgG and anti-H3K9me3, respectively, and nxf2 promoter-specific PCR primers in triplicates as indicated in Fig. 5B. The H3K9me3 was normalized to input. G. CD8+ CTLs were isolated from pooled tumor tissues of five CT26 tumor-bearing mice from the control and F5446 treatment groups as shown in Fig. 6E. The isolated CTLs were also analyzed by qPCR in triplicates for the expression of genes for granzyme B, perforin, FasL, and IFNγ. Differences in each gene expression between control and the treatment groups were determined by two-tail t test with p<0.05 as being statistically significant. Column: mean; Bar, SEM.