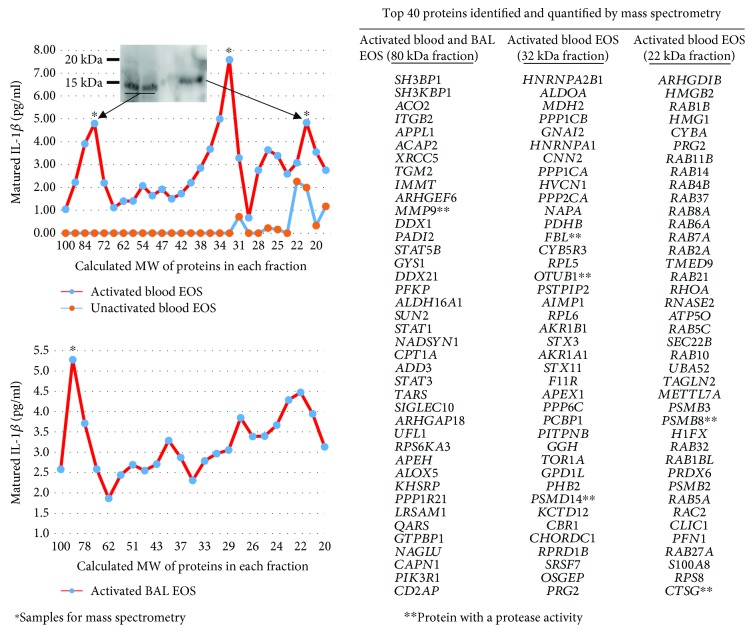
Figure 3.
Identification of matrix metalloproteinase-9 (MMP-9) as a potential protease for IL-1β cleavage in cytokine-activated eosinophils. Blood and BAL eosinophils were cultured in medium alone (inactivated) or with TNF-α plus IL-3 (both at 10 ng/ml; activated) for 48 h and 72 h, respectively. Using a zymography approach, eosinophil lysates were electrophoresed in a 12% SDS-PAGE gel copolymerized with pro-IL-1β. The pro-IL-1β zymograms were renatured and incubated overnight under conditions compatible with the activation of proteases. Each lane of the pro-IL-1β zymogram was excised and sequentially cut into 1 mm slices. The sizes of proteins (MW) in each gel slice are shown on the x-axis of the graphs. Proteins were allowed to diffuse out of the gel slices. The y-axis of the graphs indicates the amount of matured/cleaved IL-1β present in each gel slice as measured by ELISA. As shown on the upper graph, gel slices underwent western blotting for IL-1β to identify the size of the IL-1β proteins. Western blot using the ~80 kDa gel slice from two experiments using blood eosinophils from two different subjects is shown, and it identifies the IL-1β with a size of ~15 kDa. The ~80 kDa, ~32 kDa, and ~22 kDa (∗) gel slices were also submitted to the University of Wisconsin Biotechnology Center for mass spectrometry-based proteomics. On the right side of the figure are listed the top 40 genes coding for identified proteins. The lists of genes coding for proteins present in (i) the ~80 kDa gel slice for both activated blood and BAL eosinophils (n = 2), (ii) the ~32 kDa gel slice in activated blood eosinophils (n = 2), and (iii) the ~22 kDa gel slice in activated eosinophils (n = 1) are shown. Genes with asterisk possess a protease activity according to DAVID Bioinformatic Resources 6.8 (beta) (National Institute of Allergy and Infectious Diseases (NIAID), NIH) [65].