Dimethyl fumarate (DMF) is a reactive fumarate ester used in the treatment of relapsing remitting multiple sclerosis; however, the neuroprotective mechanisms of DMF action are incompletely understood. The results uncover novel DMF-modified cysteine residues in neurons and astrocytes, including cytoskeletal proteins whose modulation by DMF may alter the response to neurodegenerative cues and myelination.
Keywords: Chemical biology, Drug targets*, Post-translational modifications*, Protein adducts, Mechanism of action, Dimethyl fumarate, Succination
Graphical Abstract
Highlights
Dimethyl fumarate covalently modifies cysteine residues in neurons and astrocytes.
Cofilin-1, tubulin and collapsin response mediator protein 2 (CRMP2) are targets.
DMF-modified cofilin-1 reduces actin-severing ability, preserving filamentous actin.
Abstract
The fumarate ester dimethyl fumarate (DMF) has been introduced recently as a treatment for relapsing remitting multiple sclerosis (RRMS), a chronic inflammatory condition that results in neuronal demyelination and axonal loss. DMF is known to act by depleting intracellular glutathione and modifying thiols on Keap1 protein, resulting in the stabilization of the transcription factor Nrf2, which in turn induces the expression of antioxidant response element genes. We have previously shown that DMF reacts with a wide range of protein thiols, suggesting that the complete mechanisms of action of DMF are unknown. Here, we investigated other intracellular thiol residues that may also be irreversibly modified by DMF in neurons and astrocytes. Using mass spectrometry, we identified 24 novel proteins that were modified by DMF in neurons and astrocytes, including cofilin-1, tubulin and collapsin response mediator protein 2 (CRMP2). Using an in vitro functional assay, we demonstrated that DMF-modified cofilin-1 loses its activity and generates less monomeric actin, potentially inhibiting its cytoskeletal remodeling activity, which could be beneficial in the modulation of myelination during RRMS. DMF modification of tubulin did not significantly impact axonal lysosomal trafficking. We found that the oxygen consumption rate of N1E-115 neurons and the levels of proteins related to mitochondrial energy production were only slightly affected by the highest doses of DMF, confirming that DMF treatment does not impair cellular respiratory function. In summary, our work provides new insights into the mechanisms supporting the neuroprotective and remyelination benefits associated with DMF treatment in addition to the antioxidant response by Nrf2.
Fumarate esters have been utilized for the treatment of autoimmune psoriasis for two decades (1). More recently, a dimethyl fumarate (DMF)1 formulation has been developed for the treatment of relapsing-remitting multiple sclerosis (RRMS), a chronic inflammatory condition resulting in neuronal demyelination and axonal loss (2). Two randomized, double-blind, placebo-controlled trials (DEFINE and CONFIRM) with DMF demonstrated that treatment of RRMS results in sustained clinical and neuroradiological efficacy, and a reduced progression toward disability (3–5). As a result of these positive outcomes, this DMF formulation (marketed as Tecfidera®) was approved in the US in March 2013. DMF therapy is associated with beneficial immunomodulatory and neuroprotective effects; however, the complete mechanism of action remains unknown.
In the current study we propose that DMF chemically modifies cysteine residues on a range of intracellular protein targets. Early studies demonstrated that fumarate esters mediated anti-inflammatory effects through the modulation of T helper (Th) cells (6, 7), inducing a shift from the Th1 profile toward a favorable Th2 profile and the production of IL-4 and IL-5 (8), as well as an increase in type II dendritic cells (DCs) (9). Mechanistically, the immunomodulatory and neuroprotective effects are mediated in part through the cysteine modification of both reduced glutathione and reactive thiols on Kelch-like ECH-associated protein 1 (Keap1) by the fumarate esters (10, 11). The depletion of reduced glutathione and modification of Keap1 results in the stabilization of nuclear factor (erythroid-derived 2)-related factor 2 (Nrf2), a transcription factor regulating the cellular response to oxidative stress via the transcription of antioxidant response element (ARE) genes. This leads to an increase in proteins such as heme oxygenase 1 (HO-1), NADPH-quinone-oxidoreductase-1 (NQO1) and glutamate cysteine ligase (GCL), facilitating the replenishment of glutathione and a sustained defense against oxidative stress (12, 13). Although Nrf2 activation remains the primary described mechanism of action of DMF (14–16), recent studies demonstrate that DMF is therapeutically beneficial for the treatment of multiple sclerosis models in Nrf2 knockout mice (17). Upon ingestion DMF may be rapidly converted to monomethyl fumarate (MMF, generated after removal of a methyl group), and MMF agonism of the hydroxycarboxylic acid receptor 2 (HCAR2) also appears to be responsible for some of the positive immunologic effects of DMF therapy (18). A comprehensive quantitative proteomic approach (isotopic tandem orthogonal proteolysis-activity-based protein profiling, isoTOP-ABPP), focused on the immune system has recently identified ∼40 DMF-sensitive cysteines in primary human T cells, confirming the reactivity of this potent electrophile with immunomodulatory proteins including inhibitor of κβ kinase (IKKβ), tumor necrosis factor-α-induced protein 3 (TNFAIP3) and IL-16 (19). DMF decreased cell surface levels of the IL-2 receptor to a similar level in Nrf2+/+ and Nrf2−/− mouse splenocytes, again suggesting that the modulation of T cell activation by DMF involves additional protein targets. Blewett et al. demonstrated that the modification of a CXXC motif on PKCθ by DMF disrupted its interaction with CD28 at the immunological synapse, preventing T cell activation and IL-2 production (19).
We have recently described the increased succination of protein thiols by endogenously produced fumarate in the brainstem of NDUFS4 knockout mice (20), a model of the mitochondrial disease Leigh Syndrome, prompting us to consider further the impact of succination in neurons. The chemical modification of proteins by fumarate yields S-(2-succino)cysteine (2SC) (21), and is also increased in adipocytes under diabetic conditions (22–25) as well as fumarase deficient tumors (26, 27), where fumarate also leads to the succination of Keap1 thiols (28). We have used several molecular and chemical experimental approaches to increase intracellular fumarate levels, including DMF treatment (29), and we have described the reactivity of tubulin thiols with DMF in vitro (30). When using DMF to rapidly increase protein succination in cells in vitro, we had predicted that the methyl groups of DMF would be hydrolyzed by esterases upon entry into the cell, and that fumarate would react with protein thiols and be detected using a specific anti-2SC antibody (22). However, the lack of visible protein succination upon DMF or MMF treatment led us to consider that the continued presence of the methyl groups was preventing detection of the modified proteins using the anti-2SC antibody. It is therefore significant to note that in vitro DMF treatment does not replicate the modification generated by endogenous fumarate accumulation, such as that observed in the fumarate deficient tumors described above. Consequently, we designed a brief saponification procedure to facilitate immunodetection and observed a wide range of succinated protein bands by immunoblotting (29). In the current investigation we first confirmed increased modification of proteins by DMF using this procedure, followed by a proteomic approach to identify and confirm the site of modification of novel DMF-modified proteins in neurons and astrocytes. We propose that DMF exerts direct effects in neural cell populations that are independent of the activation of the peripheral immune response to modulate the disease progression in multiple sclerosis. This is supported by evidence that DMF and its metabolites accumulate in the brain and induce differential changes in gene expression in regions such as the cortex, cerebellum and hippocampus (17, 31). We further propose that the mechanism of action of DMF in neural cell populations is not centered solely upon the activation of the Keap1/Nrf2 antioxidant system; instead other abundant targets of DMF modification may impact a broad range of cellular functions that also contribute to therapeutic efficacy. We performed select functional analyses on several of the novel protein targets we identified to determine if DMF modification altered well-characterized functions of these proteins; in some cases, the reactive thiols identified on these proteins have been studied in the context of neurodegenerative processes. These observations better define the extensive action of DMF in vitro and provide more insight on neuroprotective mechanisms that may be exploited to improve MS treatment through the design of targeted therapeutics.
EXPERIMENTAL PROCEDURES
Chemicals
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Co (St. Louis, MO). Criterion polyacrylamide gels (Cat # 567–1043) and Precision Plus protein ladder (Cat # 161–0374) were purchased from BioRad Laboratories (Richmond, CA). Polyvinylidene difluoride (PVDF) membranes (Cat # 10600023) were from GE Healthcare (Piscataway, NJ). Pierce ECL 2 Western blotting Substrate (Cat # 80196) was from Thermo Scientific (Rockford, IL). The preparation of the polyclonal anti-2SC antibody has been described previously (22). The following commercial antibodies were used: α-tubulin (clone DM1A, Cat # 3873), fumarase (clone D9C5, Cat # 4567), succinate dehydrogenase a (SDHa, clone D6J9M, Cat # 11998) and ubiquitin (clone P4D1, Cat # 3936) from Cell Signaling Technology, Inc. (Danvers, MA); α-tubulin (clone B-7, Cat # sc-5286) from Santa Cruz Biotechnology (Dallas, TX); heme oxygenase-1 (HO-1, Cat # ADI-SPA-896-F) from Enzo Life Sciences (Farmingdale, NY); glial fibrillary acidic protein (GFAP, Cat # MAB360) from EMD Millipore (Billerica, MA) and ubiquitin C-terminal hydrolase L1 (Uchl1, Cat # PAB3903) from Abnova (Taipei City, Taiwan). MitoProfile antibody mixture for the detection of electron transport chain components (Cat # ab110413) was from MitoSciences (Eugene, OR).
Primary Neuron Isolation and Culture
All animal use described in this and other sections was consistent with the guidelines issued by the National Institutes of Health and were approved by the University of South Carolina Institutional Animal Care and Use Committee. Primary neurons from newborn rat brain cortices were isolated and cultured using an adaptation of the method described by Brewer (32). Briefly, postnatal day 1 rats were sacrificed by decapitation, the brains were aseptically dissected and cortices were separated from the rest of the brain in ice-cold Hibernate A medium (Cat # A1247501, Gibco/Thermo Fisher Scientific, Waltham, MA), containing 2% (v/v) B-27 supplement (Cat # 17504044, Gibco/Thermo Fisher Scientific) and 0.5 mm glutamine (Cat # 25030149, Gibco/Thermo Fisher Scientific). The tissue was minced in fragments of about 1 mm3 with a scalpel and subjected to digestion with 2 mg/ml papain (Cat # LS003120, Worthington Biochemical Corp., Lakewood, NJ) in the supplemented Hibernate A medium for 20 min at 30 °C in a shaker incubator set at 100 rpm. After thorough trituration through a fire polished Pasteur pipette, the tissue was allowed to settle for 5 min and the supernatant was carefully layered on top of a discontinuous OptiPrep (Cat # d-1556, Sigma-Aldrich) gradient prepared in Hibernate A medium; the layers contained 35, 25, 20 and 15% OptiPrep. The gradient was centrifuged at 800 g for 15 min at room temperature, and layers 1 (15% OptiPrep) and 2 (20%) were discarded. Layer 3 (25%) was collected and added a 5-fold volume of Neurobasal A medium (Cat # 10888022, Gibco/Thermo Fisher Scientific), containing 0.5 mm glutamine and 2% B-27 supplement. After a centrifugation at 500 × g for 5 min, cells were resuspended in Neurobasal A medium containing 0.5 mm glutamine, 2% B-27 supplement, and 5 ng/ml bFGF (Cat # 13256029, Invitrogen/Thermo Fisher Scientific); counted and plated on 24-well plates pretreated with 0.01% poly-l-lysine (Cat # P4707, Sigma-Aldrich) at a density of 200,000 cells/well. Fifty percent of the media was replaced every third day, with the addition of 5 μm AraC (Cat # BP2512100, Fisher Scientific, Thermo Fisher Scientific, Waltham, MA) from DIV 3 to inhibit glial proliferation. On DIV 8, cells were left untreated or treated for 24 h with 10 μm or 100 μm dimethylfumarate (DMF, Cat # 242926, Sigma-Aldrich) prepared in Dulbecco's PBS (DPBS, Cat # 21316003, Corning Cellgro, Manassas, VA) and filtered. On DIV 9, medium was removed, cells were rinsed 3 times with DPBS and collected after the addition of 250 μl radioimmunoprecipitation assay (RIPA) lysis buffer [50 mm Tris-HCl (Cat # BP152–5, Fisher Scientific, Thermo Fisher Scientific), 150 mm NaCl (Cat # S7653, Sigma-Aldrich), 1 mm EDTA (Cat # ED2SS, Sigma-Aldrich), 0.1% Triton X-100 (Cat # T9284, Sigma-Aldrich), 0.1% SDS (Cat # BP166–5, Fisher Scientific, Thermo Fisher Scientific), 0.5% sodium deoxycholate (Cat # D6750, Sigma-Aldrich), pH 7.4], with the addition of 2 mm diethylenetriaminepentaacetic acid (Cat # D6518, Sigma-Aldrich), and a protease inhibitor mixture (Cat # P8340, Sigma-Aldrich). Homogenization was performed by pulse sonication at 2 watts using a Model 100 sonic dismembrator (Thermo Fisher Scientific) for 30 s before resting on ice for 30 min in lysis buffer as described before (30). Protein in the lysates was precipitated with 9 volumes of cold acetone for 10 min on ice. After centrifugation at 3000 × g for 10 min and removal of the acetone, the protein pellet was resuspended in 150 ìl RIPA buffer. The protein content in the lysates was determined by the Lowry assay (33).
Primary Astrocyte Isolation and Culture
Primary astrocytes were isolated and cultured from neonatal P1 - P2 Sprague-Dawley rats. Animals were decapitated following which cortices, brainstem, and hippocampi were aseptically dissected in Petri dishes filled with ice-cold Hank's Balanced Salt Solution (HBSS; Cat # 14025–076, Gibco, Thermo Fisher Scientific), HEPES (10 mm; Cat # H0753, Sigma-Aldrich), and gentamicin sulfate (5 μg/ml; Cat # 15710064, Gibco, Thermo Fisher Scientific). The dissected tissue was cut into pieces, and dissociated by trypsin treatment (10 μl/ml; Cat # 27250018, Gibco/Thermo Fisher Scientific), followed by trituration with sterile glass pipettes. Cells were plated in 24-well plates precoated with poly-l-lysine (50 μg/ml). Cultures were maintained in DMEM/F12 (Cat # 11320–033, Gibco/Thermo Fisher Scientific) medium supplemented with 10% fetal bovine serum (FBS, Cat # S11150, Atlanta Biologicals Inc., Flowery Branch, GA), and stored in an incubator at 37 °C (5% O2/95% CO2). At day in vitro 1 (DIV1), cells received a full medium replacement with subsequent half medium replacements every third day. This protocol results in astrocyte-enriched cultures as confirmed by immunostaining against the astrocytic marker, glial fibrillary acidic protein (GFAP, supplemental Fig. S2). Cells were treated with 10–100 μm DMF for 24 h and harvested at DIV10–12 as described above for primary neurons.
N1E-115 Cell Culture
N1E-115 cells (subclone N1E-115–1 neuroblastoma cells) were obtained from Sigma-Aldrich (Cat # 08062511). The cells were expanded in nondifferentiation medium (NDM): 90% DMEM (Cat # 12430–054, Gibco/Thermo Fisher Scientific, containing with 25 mm glucose, no pyruvate, 25 mm HEPES, 4 mm glutamine) and 10% FBS. At 80% confluence, the cells were differentiated into neurons in the presence of 2% FBS and 1.25% dimethyl sulfoxide (DMSO, Cat # 32434, Alfa Aesar, Thermo Fisher Scientific) in DMEM for 5 days (34). In addition to the assessment of the neuronal phenotype by light microscopy, the detection of synaptophysin protein levels confirmed successful differentiation. During the final 24 h of differentiation the cells were treated with 10–100 μm DMF and protein was collected as described for primary neurons.
Lentiviral Transduction of 3T3-L1 Fibroblasts
The lentiviral vectors were prepared by the University of South Carolina Viral Vector Facility. Briefly, TRC2 Fh1 shRNA, clone- TRCN0000246831 or MISSION TRC2 pLKO.5-puro nonmammalian shRNA control plasmids (Cat # SHC202, Sigma-Aldrich) were used to generate the lentiviral vectors that also contained a puromycin resistance gene. Fifteen microgram vector plasmid, 10 μg psPAX2 packaging plasmid (Cat # 12260, Addgene, Cambridge, MA), 5 μg pMD2.G envelope plasmid (Cat # 12259, Addgene) and 2.5 μg pRSV-Rev plasmid (Cat # 12253, Addgene) were transfected into 293T cells. The filtered conditioned medium was collected and stored at −80 °C. 3T3-L1 fibroblasts (Cat # CL-173™, ATCC, Manassas, VA) were incubated overnight with 150 μl of filtered conditioned medium containing Fh1 shRNA or control lentivirus. Successfully transduced fibroblasts were selected using 1 μg/ml puromycin (Cat # P9620, Sigma-Aldrich). The selected fibroblasts were propagated in the presence of puromycin until confluent and harvested in RIPA buffer as described above. Successful knockdown of fumarase expression was determined by immunoblotting and fumarate levels were determined by GC-MS as described previously (20).
Saponification of Fumarate Esters
Sixty micrograms of protein from control and DMF treated cell lysates was incubated with 80% DMSO, 6 mm potassium hydroxide (KOH, Cat # 484016, Sigma-Aldrich), and 1 mm EDTA at room temperature for 30 min, with vortexing at 5 min intervals. The pH was adjusted to 7, and the protein was precipitated with 90% acetone (Cat # BDH1101–4LG, VWR, Radnor, PA) before being resuspended in 40 μl RIPA buffer. The pH was again adjusted to 7 before gel electrophoresis and immunoblotting.
One-dimensional PAGE and Western Blotting
Western blotting was performed as described previously, after separation of the proteins by SDS-PAGE (22, 23). For protein identification purposes gels were stained with Coomassie Brilliant Blue R (Cat # 27816, Sigma-Aldrich) following electrophoresis to allow band isolation and mass spectrometry (see below). In some cases, membranes were stripped with 62.5 mm Tris, pH 6.8, containing 2% SDS and 0.7% 2-mercapto ethanol (Cat # M6250, Sigma-Aldrich) for 20 min at 65 °C before reprobing with a different antibody.
Protein Identification from SDS-PAGE Gel Bands by LC-MS/MS
To identify the sites of fumarate ester modification, 60 μg of protein from primary neurons, 200 μg of protein from primary astrocytes or 120 μg of protein from differentiated N1E-115 neurons were resolved by SDS-PAGE, and the gels were stained with Coomassie Brilliant Blue R. The visible protein bands were excised from the gels and subjected to in-gel digestion with trypsin (the gel was cut into segments based on the intensity of the Coomassie stain and the protein bands were excised). Briefly, after destaining the proteins were reduced with 10 mm dithiothreitol (Cat # V3155, Promega, Madison, WI) and alkylated with 170 mm 4-vinylpyridine (Cat # V3204, Sigma-Aldrich). Protein digestion was carried out overnight at 37 °C in the presence of 500 ng sequencing grade modified trypsin (which cleaves C-terminal to lysine and arginine residues, Cat # V5280, Promega) in 50 mm ammonium bicarbonate (Cat # 09830, Sigma-Aldrich). After gel extraction, the peptides were analyzed in a blinded manner on a Dionex Ultimate 3000-LC system (Thermo Scientific) coupled to a Velos Pro Orbitrap mass spectrometer (Thermo Scientific). The LC solvents were 2% acetonitrile (Cat # 34851, Sigma-Aldrich)/0.1% formic acid (Cat # 85178, Pierce, Thermo Scientific) (Solvent A) and 80% acetonitrile/0.1% formic acid (Solvent B); the water used for these solvents was LC-MS grade (Cat # 39253, Honeywell International Inc., Morris Plains, NJ). Peptides were first trapped on a 2 cm Acclaim PepMap-100 column (Thermo Scientific) with Solvent A at 3 μl/min. At 4 min the trap column was placed in line with the analytical column, a 75 μm C18 stationary-phase LC PicoChip Nanospray column (New Objective, Inc., Woburn, MA). The peptides were eluted with a gradient from 98%A:2%B to 40%A:60%B over 30 min, followed by a 5 min ramp to 10%A:90%B that was held for 10 min. The Orbitrap was operated in data-dependent acquisition MS/MS analysis mode and excluded all ions below 200 counts. Following a survey scan (MS1, up to 8 precursor ions were selected for MS/MS analysis. All spectra were obtained in the Orbitrap at 7500 resolution. The DDA data were analyzed using Proteome Discover 1.4 software with SEQUEST algorithm against the uniprot_ref_mouse database (2014–10−03 version, 52,474 proteins) or uniprot_ref_rat database (2011–5-11 version, 39765 proteins) with XCorr validation >2 (+2) or >2.5 (+3). An allowance was made for 2 missed cleavages following trypsin digestion. No fixed modifications were considered. The variable modifications of methionine oxidation (MOX), proline hydroxylation (POX), cysteine pyridylethylation (CPE, 105.058) or cysteine succination by fumarate (C2SC, 116.011), cysteine modification by monomethyl fumarate (CMMF, 130.026) or cysteine modification by dimethyl fumarate (CDMF, 144.042) were considered with a mass tolerance of 15 ppm for precursor ions and a mass tolerance of 10 ppm for fragment ions. The results were filtered with a false discovery rate of 0.01 for both proteins and peptides (Percolator node). A minimum of 8 unique peptides was reported for all proteins identified (maximum 106 peptides). For all identifications the spectra were manually inspected to confirm identity of the proposed DMF-modified Cys containing peptides. Any low-quality spectra/incorrect identifications were discarded before performing MS/MS analyses on select masses of interest from the high quality protein identifications.
Select MS/MS Analyses
The samples were reanalyzed to target the expected DMF-modified Cys containing peptide mass from the data obtained in DDA mode, further MS/MS analyses were used to monitor select modified peptide masses of interest. The spectrometer repeatedly acquired an MS1 spectrum followed by the desired MS/MS spectra (CID fragmentation in the ion trap, all spectra in the Orbitrap at 7500 resolution). These MS/MS spectra were then averaged over the entire LC peak yielding a spectrum superior to that obtained by DDA (where the acquisition time was split looking for many peptides at once). The CID-MS/MS data was inspected in Proteome Discover 1.4 software for the masses of interest with either cysteine modification by monomethyl fumarate (CMMF, 130.026), or cysteine modification by dimethyl fumarate (CDMF, 144.042) being considered based on the expected modification from the DDA run. The variable modifications of methionine oxidation (MOX), proline hydroxylation (POX) or cysteine pyridylethylation (CPE, 105.058) were considered. The MS/MS spectra were inspected again using Thermo Xcalibur 2.2 software. Manual sequencing of the spectra was used to confirm the sequence and modification site of the peptides. The mass spectrometry analyses constitute a Tier 3 measurement and the proteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE (35) partner repository with the data set identifier PXD008314. The file names in the PRIDE data reflect the protein identified in the original DDA run from a gel band.
DRG Culture and Fluorescence Time-Lapse Microscopy
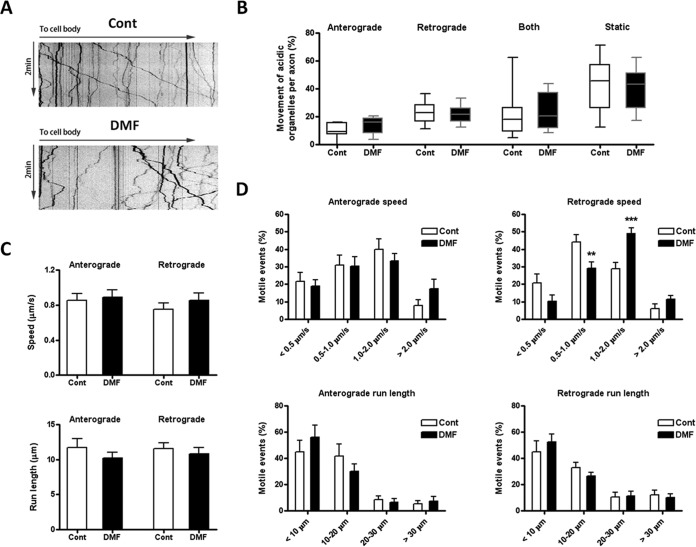
Dorsal Root Ganglion (DRG) neurons were isolated from rat sciatic nerve as described previously (36). 48 h after sciatic nerve crush to induce a regenerative response, adult rat DRG neurons were plated for 16–24 h onto poly-d-lysine (Cat # P6407, Sigma-Aldrich) and laminin (Cat # 11243217001, Sigma-Aldrich)-coated German glass coverslips and maintained in Ham's F12 medium (Cat # 11765–047, Gibco, Thermo Fisher Scientific) supplemented with 10% horse serum (Cat # 16050–122, Gibco, Thermo Fisher Scientific). Neurons were exposed to 500 nm Lysotracker Red (Cat # L7528, Invitrogen, Thermo Fisher Scientific) for 5 min before imaging. Coverslips were transferred into fresh medium containing 25 mm HEPES (Cat # 15630080, Gibco, Thermo Fisher Scientific), pH 7.4 and 1% OxyFluor (Cat # OF-0005, Oxyrase, Inc., Mansfield, OH) in a water-heated microscope stage warmed to 37 °C. Cells expressing a relatively low level of Lysotracker Red were selected for time-lapse imaging using an Axiovert 200 inverted microscope (Carl Zeiss, Inc.). Fluorescent images were acquired every 0.3 s for 2 min using a Plan-Apo 63×/1.2 W/0.17 water objective. Kymographs were generated from time-lapse movies using NIH ImageJ software. The kymographs were generated such that the direction toward the cell body was always to the right, so lines that sloped toward the right at any point with a net displacement of >5 μm were categorized as retrograde organelles. Lines that sloped toward the left >5 μm at any time during the recording interval were considered anterograde organelles. Lines that zigzagged were categorized as bidirectional, and lines that showed <5 μm lateral displacements in any direction during the recording interval were categorized as static. A total of fifteen axons per experimental group in three independent experiments were used for quantification purposes, and the number of organelles for each category of movement expressed as % of total organelles was used to construct a box and whiskers graph (Fig. 4B). The data was further analyzed by calculating the speed (in μm/s) and run length (in μm) for the anterograde and retrograde motile events; results were expressed as mean ± S.E. (Fig. 4C). Finally, the speed and run length for anterograde and retrograde motile events were grouped in 4 different intervals, transformed in % and expressed as mean ± S.E. (Fig. 4D).
Fig. 4.
DMF effects on lysosomal motility in rat dorsal root ganglion (DRG) neurons. Rat DRG neurons were plated on coverslips, treated with PBS (control) or 50 μm DMF for 24 h, followed by 500 nm Lysotracker for 5 min to stain acidic organelles. Lysosome movements were recorded for 2 min with 0.3 s intervals, as described under Experimental Procedures. A, Representative acidic organelle kymographs in neurons from control or 50 μm DMF treated groups. B, The percentage of organelles showing (1) anterograde movement (away from the cell body), (2) retrograde movement (toward the cell body), (3) both movements (switching directions one or more times), and (4) static behavior (no movement) were determined for 15 axons in three different experiments. Data is presented as a box and whiskers graph; no differences were observed because of the DMF treatment for each movement, as determined by one-way ANOVA and Tukey's multiple comparisons post-test. C, Speed and run length of both anterograde and retrograde motile events. Results were expressed as mean ± S.E.; no differences were observed by unpaired Student's t test. D, DMF treatment changed the distribution of speed in retrograde motile events (upper right panel). Compared with control, there was higher percentage of 1–2 μm/s motile events but lower percentage of 0.5–1 μm/s motile events in the DMF treated group. No changes in the distribution of speed in anterograde motile events (upper left panel), run length in anterograde motile events (lower left panel) and run length in retrograde motile events (lower right panel) were observed. Results were expressed as mean ± S.E., and significance was determined by one-way ANOVA and Tukey's multiple comparisons post-test. *p < 0.05 and ***p < 0.001 for DMF treated versus control neurons.
Cofilin Activity Assay
Cofilin activity was determined by measuring its ability to sever actin filaments, as described by Yonezawa et al. (37). Briefly, aliquots of 7.5 μg human recombinant cofilin (containing Tris, NaCl, sucrose and dextran, Cat # CF01, Cytoskeleton Inc., Denver, CO) were dissolved in water (final concentrations:10 mm Tris-HCl, pH 8.0, 10 mm NaCl, 5% sucrose, 1% dextran), added sodium diethylenetriaminepentaacetate (Cat # D6518, Sigma-Aldrich) to 100 μm, and Tris(2-carboxyethyl)phosphine hydrochloride (Cat # 20490, Thermo Fisher Scientific) to 250 μm, and incubated overnight at room temperature with a 5-fold molar excess of dimethylfumarate (DMF), or with vehicle. The following day, aliquots of 15 μg rabbit muscle actin (Cat # AKL99, Cytoskeleton Inc.) dissolved in ice-cold General Actin Buffer (5 mm Tris-HCl pH 8.0, 0.2 mm CaCl2, Cat # BSA01001, Cytoskeleton Inc.) supplemented with 0.2 mm ATP (Cat # BSA04, Cytoskeleton Inc.), were polymerized into F-actin by addition of 1/10 volume of Actin Polymerization Buffer (100 mm Tris-HCl pH 7.5 containing 20 mm MgCl2, 500 mm KCl, 10 mm ATP and 50 mm guanidine carbonate, Cat # BSA02, Cytoskeleton Inc.) followed by 1 h incubation at room temperature. At this point, the pH of the F-actin aliquots was adjusted to 8.0 with an excess of 10 mm Tris-HCl before the addition of the cofilin aliquots that were incubated overnight with or without DMF, and the incubation continued for another 30 min at room temperature. The samples were then centrifuged at 150,000 × g for 1 h at 25 °C, with deceleration set without brakes. After separation of the supernatants (containing depolymerized actin monomers) from the pellets (containing F-actin), fractions of supernatants and pellets containing the equivalent to 3 μg actin and 1.5 μg cofilin in the initial mix were resolved by SDS-PAGE, and gels were stained with Coomassie Brilliant Blue R (n = 4 replicates per experimental group). Images of the gels were captured using a Gel Doc XR+ scanner (Bio-Rad Laboratories, Inc.), and the integrated OD x area value of the bands was analyzed with the Image Lab Software V5.2.1 (Bio-Rad Laboratories, Inc.). For each particular protein (actin and cofilin), results of the band integrated OD in supernatants and pellets were expressed as % of the total integrated OD (supernatant band + pellet band), grouped by treatment, and then reported as the mean ± S.E. Additional control experiments included F-actin samples that did not have added cofilin, or F-actin samples that were adjusted to pH 6.8 (to inhibit cofilin severing activity on F-actin) before cofilin addition.
Ubiquitin C-Terminal hydrolase L1 (Uchl1) Activity Assay
UchL1 deubiquitinase activity was assessed using recombinant UchL1 protein (Cat # 50690-M07E, Sino Biological, Beijing, China) and the substrate Ubiquitin-Rhodamine110-Glycine (Cat # U-555, BostonBiochem, Cambridge, MA). UchL1 protein (0.1 μg) was preincubated with 0, 25, 50, or 100 μm DMF for 24 h prior the addition of 0.1 μm fluorogenic ubiquitin rhodamine in a deubiquitination buffer (50 mm Tris-HCl, 150 mm NaCl, 5 mm DTT). Fluorescence was measured at 535 nm (following excitation at 485 nm) over a 2 h period using the Tecan SAFIRE Spectrofluorimeter. The initial reaction velocities were analyzed and the results were averaged for each experimental group and expressed as the mean ± S.E. n = 5 per condition analyzed).
Measurement of Oxygen Consumption Rate (OCR)
N1E-115 cells were seeded on V7 cell culture microplates coated with 0.2% gelatin (Cat # G1890, Sigma-Aldrich) at a density of 10,000 cells/well. After 3 days in culture, the cells were differentiated for 5 days as described above, and treated with 0, 10, or 50 μm DMF for the last 24 h (n = 6/group). The Seahorse extracellular flux analyzer XF-24 (Agilent Technologies, Inc., Santa Clara, CA), was used to measure the oxygen consumption rate (OCR), using XF Assay Medium (Cat # 102365–100, Agilent Technologies, Inc.) supplemented with 25 mm glucose (Cat # G8270, Sigma-Aldrich) (38). After measurement of basal respiration, oligomycin (5 μg/ml, Cat # O4876, Sigma-Aldrich), carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, 0.5 μm, Cat # C2920, Sigma-Aldrich), and rotenone (3 μm, Cat # R8875, Sigma-Aldrich) + antimycin A (4 μm, Cat # A8674, Sigma-Aldrich) were added sequentially to determine ATP production/proton leak, spare respiratory capacity and nonmitochondrial respiration. After completion of the assay, the medium was removed and the cells were immediately washed 3 times with cold PBS. The plate was stored at −70 °C before the measurement of the total protein content to normalize individual measurements. The results were averaged for each experimental group and expressed as the mean ± S.E.
Experimental Design and Rationale
The purpose of these experiments was to identify novel targets of DMF mediated electrophilic modification of cysteine residues in neurons and astrocytes in vitro, in an attempt to better understand the biological targets and mechanism of action of this drug. The detection of fumarate ester derived succination will only be present in the DMF treated cells. For experiments using primary rodent cultures, at least 3–6 rodent pups were used to generate cells for primary culture. A minimum of three replicate treatment plates/wells (untreated controls versus DMF treated) were used and each separate experiment was reproduced with new cells one-four times. In some cases, immunoblotting for increased heme oxygenase −1 was used to confirm a biological effect of DMF treatment on the cells. Protein from individual cell culture replicates were separated by electrophoresis and peptides extracted from parallel excised gel bands were pooled in order to obtain enough material for confirmation of the site of modification. At least three technical repeats (LC-MS/MS analyses) were conducted in order to confirm the protein identification and site of modification. All other analyses (including respiration analyses and monitoring of lysosomal trafficking) were performed with a minimum of 3 independent biological replicates per group, with some up to 8 replicates per group (n = 3–8). The Uchl1 deubiquitinase activity was studied with n = 5 per group. Data are summarized throughout as mean ± S.E. and are plotted using SigmaPlot 11 software (Systat Software, Inc. San Jose, CA) and Prism 4 (GraphPad Software, La Jolla, CA). Statistical analyses were performed using SigmaPlot 11 and Prism 4. Differences between more than two groups were analyzed using one-way ANOVA with either the Holm- Sidak or Tukey's post-test. When two groups were compared, the unpaired Student “t” test was used. In all cases, p < 0.05 was considered statistically significant.
RESULTS
DMF-Induced Protein Modification in Neural Cells
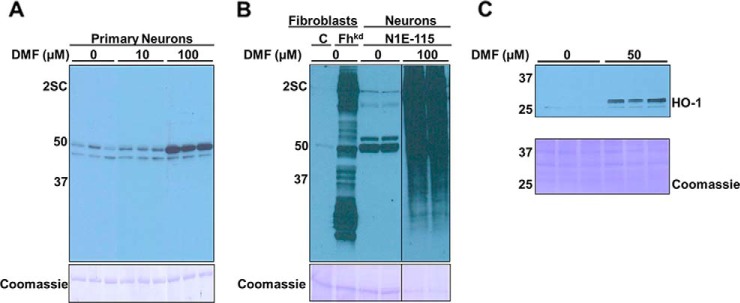
Although the immunomodulatory and neuronal benefits of DMF are often attributed to the succination of Keap1 and activation of the antioxidant response element (ARE), we predicted that a wider range of protein thiols might also be modified by DMF or its primary metabolite monomethyl fumarate (MMF), because we had observed that this occurs in adipocytes in vitro (29). In order to detect succinated proteins in neurons treated with DMF for 24 h, we employed a procedure that we had developed previously using alkaline hydrolysis to remove the ester and permit the immunological detection of protein succination (supplemental Fig. S1B) (29). In the absence of ester hydrolysis there was limited detection of succinated proteins in rat primary neurons using the anti-2SC antibody, with only one band ∼50 kDa showing a significant increase in intensity following 100 μm DMF treatment (Fig. 1A, lanes 7–9). This indicated that at least one or both of the methyl groups had not been removed by intracellular esterases and was preventing interaction with the anti-2SC antibody that recognizes the S-(2-succino)cysteine epitope (22). To ensure that the succinated proteins detected were solely of neuronal origin we differentiated N1E-115 neuroblastoma cells to a neuronal phenotype (confirmed by increased synaptophysin protein content) (20), and treated these with DMF for 24 h. The hydrolysis of the ester in the presence of KOH facilitated the detection of a large number of succinated proteins (Fig. 1B, last panel, lanes 5 and 6), similar to what we have observed previously in adipocytes following fumarate ester treatment (29). Fibroblasts that had fumarase knocked down using a lentiviral mediated shRNA approach to increase endogenous fumarate levels (39) were used as a positive control for succination (Fig. 1B, lane 2). The intensity of succinated proteins in the DMF treated neurons versus the positive control indicated that fumarate esters readily enter cells and react with a wide range of protein thiols. Because DMF is known to modify thiols in Keap1 leading to the induction of ARE derived proteins such as heme oxygenase 1 (HO-1), we next confirmed that DMF treatment significantly induced HO-1 protein levels (Fig. 1C).
Fig. 1.
DMF treatment increases protein succination in rat primary neurons and in the N1E-115 neuronal cell line. A, Total cell lysates (10 μg protein) from rat primary neurons (DIV 8) treated for 24 h with vehicle (0) or 10 or 100 μm DMF were separated by SDS/PAGE without prior saponification to remove the ester groups, and succinated proteins were detected using a polyclonal anti-2SC antibody, as described under “Experimental procedures”. Succination was detected for only two bands of ∼50 and ∼45 kDa, with increased intensity of the ∼50 kDa band after 100 μm DMF treatment (2SC panel). B, After saponification, lysates from 100 μm DMF treated N1E-115 cells (lanes 5–6) show much more abundant protein succination than primary neurons in (A), confirming that removal of the ester groups increases detectability of DMF modified proteins (2SC panel). Fibroblasts in which fumarase expression was knocked-down (Fhkd) serve as a positive control for high protein succination (lane 2, 2SC panel). All the lanes were run in the same gel and transferred to the same membrane, which was later probed, and the image was cropped to show the 2SC and Coomassie panels. C, Increased levels of heme oxygenase 1 (HO-1) in N1E-115 neurons treated with 50 μm DMF treatment (versus controls). In all panels, molecular masses of marker proteins are indicated on the left-hand side. Loading controls are also shown (Coomassie panels).
In order to identify the novel protein targets in DMF treated neurons we used LC-MS/MS to analyze distinct gel bands representing electrophoretically separated proteins. The peptides obtained were analyzed to determine if modification of protein thiols by either DMF (CDMF), or the de-methylated metabolites MMF (CMMF) and fumarate (C2SC) had occurred. This proteomic approach allows targeted confirmation of the exact sites of modification and it does not require the base hydrolysis of the ester that can also result in a partial loss of protein. As noted in Table I, all of the proteins identified were modified by DMF or MMF, rather than 2SC, suggesting that both of these fumarate esters reacted with intracellular proteins more rapidly than they could be converted to fumarate. In primary rat neurons we confirmed the identity of 4 modified proteins, and in N1E-115 neurons we confirmed a total of 15 protein subunits modified by either DMF or MMF. Because the N1E-115 cultures were devoid of any glial cells that may be present in primary neuronal cultures, the identified proteins reflect true neuronal targets of fumarate ester modification. Because fumarate esters were previously shown to induce Nrf2 in glial cells in vivo (13), we also investigated additional protein targets of DMF-mediated protein modification in primary astrocyte cultures (supplemental Fig. S2A confirms the enrichment of Glial Fibrillary Acid Protein (GFAP) positive cells). Table I confirms the detection of 11 modified protein subunits in primary rat astrocytes after a 24 h treatment with up to 100 μm DMF. Overall, this targeted approach confirmed the novel identification of 24 distinct protein subunits in both neurons and astrocytes that are directly modified by either DMF or MMF.
Table I. MS/MS peptide identification for DMF-treated neurons and astrocytes. Raw data were searched with the Sequest node of Proteome Discover 1.4, using the uniprot_ref_mouse or uniprot_ref_rat databases. The variable modifications of methionine oxidation (MOX), proline hydroxylation (Pox), cysteine pyridylethylation (CPE, 105.058), cysteine succination by fumarate (C2SC, 116.011), cysteine modification by monomethyl fumarate (CMMF, 130.026) or cysteine modification by dimethyl fumarate (CDMF, 144.042) were considered.
| Protein name [uniprot name] | Protein sequence % coverage | # Unique peptides | Modified sequence | Modification site | Modified peptide PSMs | XCorr | m/z | Charge | MH+[Da] |
|---|---|---|---|---|---|---|---|---|---|
| Primary Neurons | |||||||||
| Collapsin Response Mediator Protein 2 [DPYL2] | 74.48 | 21 | GLYDGPVCDMFEVSVTPK | Cys 504 | 2 | 1.9 | 854.4096 | 2 | 1707.812059 |
| Vimentin [VIME] | 94.85 | 49 | QVQSLTCDMFEVDALK | Cys 328 | 3 | 2.42 | 789.3899 | 2 | 1577.772631 |
| Guanine nucleotide-binding protein G(o) subunit alpha [GNAO] | 55.93 | 13 | MOXVCDMFDVVSR | Cys 108 | 1 | 2.56 | 534.7373 | 2 | 1068.467333 |
| Annexin A1 [ANXA1] | 63.87 | 19 | QACDMFYIEK | Cys 12 | 2 | 1.21 | 499.7269 | 2 | 998.4465808 |
| Primary Astrocytes | |||||||||
| Filamin alpha [C0JPT7] | 59.49 | 106 | YTP CMMFGAGSYTIMVLFADQATPTSPIR | Cys 841 | 1 | 1.85 | 969.4722 | 3 | 2906.402134 |
| Myosin-9 [MYH9] | 34.93 | 39 | CMMFNGVLEGIR | Cys 694 | 10 | 1.21 | 545.7675 | 2 | 1090.524828 |
| Heat shock protein HSP 90-alpha [HS90A] | 69.3 | 21 | VFIMOXDNCDMFEELIPERYLNFIR | Cys 375 | 3 | 2.24 | 840.0648 | 3 | 2518.180088 |
| Heat shock protein HSP 90-beta [HS90B] | 67.4 | 28 | VFIMOXDSCDMFDELIPEYLNFIR | Cys 366 | 4 | 1.49 | 1239.089 | 2 | 2477.171678 |
| Collapsin Response Mediator Protein 2 [DPYL2] | 76.22 | 34 | GLYDGPVCMMFEVSVTPK | Cys 504 | 2 | 3.08 | 847.402 | 2 | 1693.796801 |
| GLYDGPVCDMFEVSVTPK | Cys 504 | 3 | 1.83 | 854.4108 | 2 | 1707.814501 | |||
| Vimentin [VIME] | 91.85 | 50 | QVQSLTCDMFEVDALK | Cys 328 | 3 | 2.25 | 789.3909 | 2 | 1577.774706 |
| QVQSLTCMMFEVDALK | Cys 328 | 3 | 1.65 | 782.3807 | 2 | 1563.754198 | |||
| Glial fibrillary acidic protein [GFAP] | 88.37 | 40 | QLQALTCMMFDLESLR | Cys 292 | 1 | 1.37 | 810.4024 | 2 | 1619.797533 |
| Annexin A1 [ANXA1] | 69.94 | 22 | GDRCDMFEDMOXSVNQDLADTDAR | Cys 189 | 1 | 2.04 | 757.6424 | 3 | 2270.912815 |
| QACDMFYIEK | Cys 12 | 1 | 1.06 | 499.7257 | 2 | 998.4441394 | |||
| Cofilin-1 [COF1] | 93.37 | 10 | HELQANCMMFYEEVKDR | Cys 139 | 1 | 4.86 | 621.9409 | 3 | 1863.808201 |
| Peptidyl-prolyl cis-trans isomerase A [PPIA] | 73.17 | 9 | VCDMFFELFADK | Cys 21 | 1 | 1.95 | 608.2839 | 2 | 1215.560594 |
| Fatty acid-binding protein, brain [FABP7] | 78.03 | 8 | TQCDMFTFK | Cys 56 | 3 | 2 | 436.1958 | 2 | 871.3869494 |
| N1E-115 Neurons | |||||||||
| ATP-citrate synthase [ACLY] | 51.77 | 39 | YICMMFTTSAIQNR | Cys 20 | 2 | 1.57 | 700.3239 | 2 | 1399.640551 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 [UCHL1] | 58.3 | 13 | NEAIQAAHDSVAQEGQCMMFR | Cys 152 | 6 | 5.87 | 686.3021 | 3 | 2056.891819 |
| NEAIQAAHDSVAQEGQCDMFR | Cys 152 | 4 | 4.54 | 690.9748 | 3 | 2070.910007 | |||
| Isoform Mt-VDAC1 of Voltage-dependent anion-selective channel protein 1 [VDAC1] | 88.69 | 17 | YQVDPDACMMFFSAK | Cys 245 | 11 | 1.67 | 737.3118 | 2 | 1473.610765 |
| T-complex protein 1 subunit beta [TCPB] | 69.35 | 24 | HGINCMMFFINR | Cys 289 | 2 | 1.03 | 602.2833 | 2 | 1203.559374 |
| T-complex protein 1 subunit alpha [TCPA] | 54.5 | 19 | LACMMFKEAVR | Cys 125 | 3 | 1.16 | 510.2606 | 2 | 1019.514086 |
| Elongation factor 2 [EF2] | 52.91 | 37 | STLTDSLVCMMFK | Cys 41 | 1 | 1.95 | 508.7895 | 2 | 1196.571825 |
| Heat shock protein HSP 90-beta [HS90B] | 76.93 | 32 | VFIMOXDSCMMFDELIPEYLNFIR | Cys 366 | 2 | 1.16 | 1232.073 | 2 | 2463.138719 |
| VFIMOXDSCDMFDELIPEYLNFIR | Cys 366 | 3 | 1.98 | 826.3831 | 3 | 2477.164843 | |||
| Heat shock protein HSP 90-alpha [HS90A] | 65.48 | 27 | VFIMOXDNCDMFEELIPEYLNFIR | Cys 375 | 5 | 2.77 | 840.0639 | 3 | 2518.177158 |
| Glyceraldehyde-3-phosphate dehydrogenase [G3P] | 72.07 | 8 | AAICMMFSGK | Cys 22 | 3 | 1.37 | 390.1814 | 2 | 779.3555163 |
| IgE-binding protein [IGEB] | 46.68 | 18 | QQCMMFAER | Cys 187 | 1 | 1.4 | 432.678 | 2 | 864.3488635 |
| LQGPPYAESPPCMMFVVR | Cys 181 | 2 | 2.09 | 871.9227 | 2 | 1742.838182 | |||
| Transgelin-2 [TAGL2] | 71.86 | 14 | NMOXACMMFVQR | Cys 124 | 2 | 2.24 | 484.2015 | 2 | 967.3957995 |
| Collapsin Response Mediator Protein 2 [DPYL2] | 61.89 | 17 | GLYDGPVCMMFEVSVTPK | Cys 504 | 13 | 3.45 | 846.4002 | 2 | 1693.79326 |
| Asparagine synthetase (glutamine-hydrolyzing) [ASNS] | 64.88 | 20 | ETFEDCDMFNLLPK | Cys 455 | 3 | 2.36 | 726.838 | 2 | 1452.668383 |
| Tubulin alpha-1A chain [TBA1A] | 74.94 | 12 | TIQFVDWCDMFPTGFK | Cys 347 | 2 | 2.64 | 843.3975 | 2 | 1685.787767 |
| Tubulin beta-5 chain [TBB5] | 75.23 | 5 | TAVCDMFDIPPR | Cys 354 | 1 | - | 558.6467 | 2 | 1115.5401 |
DMF Modification Affects Cofilin-1 Functionality
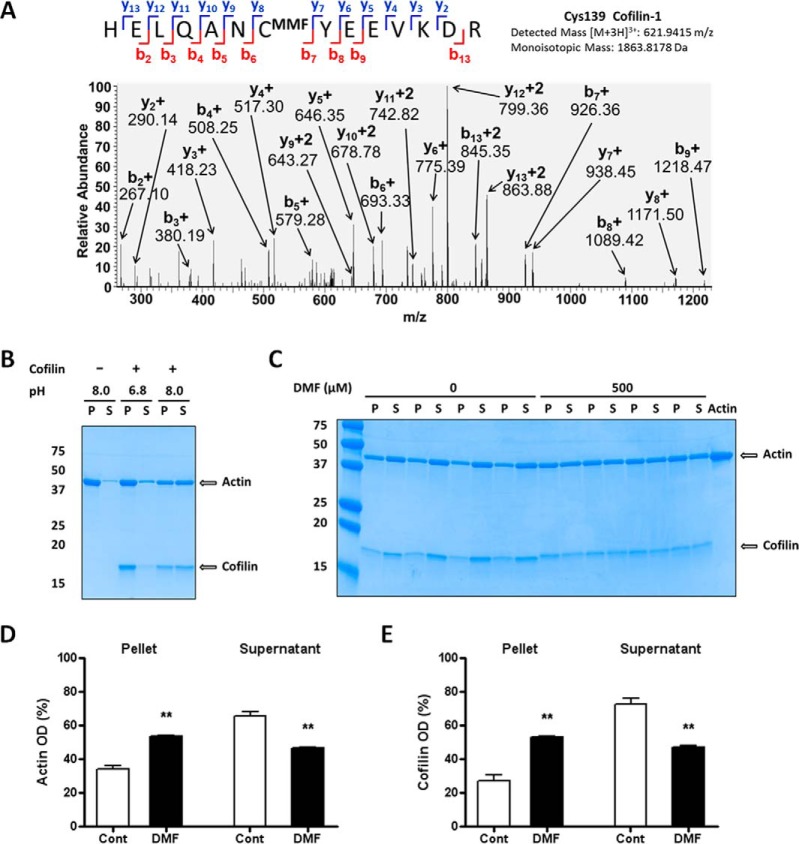
The chemical modification of several cytoskeletal proteins was observed in all cell types examined (Table I). In astrocytes this included abundant cytoskeletal proteins such as Glial Fibrillary Acid Protein (GFAP) and vimentin, as well as cofilin-1, a dynamic regulator of actin polymerization. Cofilin-1 modulates the actin cytoskeleton by depolymerizing filamentous (F) actin, generating monomeric actin that can be used to reorganize the actin cytoskeleton in response to cellular dynamics. To confirm the initial observation that Cys139 of Cofilin-1 is succinated by MMF in astrocytes in vitro, we performed additional selected reaction monitoring and detected the +3 charge state for the cofilin peptide HELQANCMMFYEEVKD (m/z 621.9415) following CID. The b7 and y8 fragment ions confirm that Cys139 is succinated by MMF (Fig. 2A). Because 100% modification of the peptide by fumarate esters was not expected, we also detected the pyridylethylated version of the same peptide (supplemental Fig. S2B, +3 charge state, 613.6218 m/z). Importantly, this cysteine residue is conserved across rat, mouse and human sequences. To determine if the succination of Cys139 impacted its activity as a regulator of actin dynamics, we examined the pH-sensitive depolymerization activity of cofilin-1 on F-actin. Cofilin binds to F-actin in a 1:1 ratio below neutral pH, but does not sever the actin filaments (37). In contrast, at pH 8.0 the presence of cofilin results in actin severing and the production of monomeric actin. Fig. 2B confirms that F-actin remains polymerized in the absence of cofilin, and in the presence of cofilin at pH 6.8 (F-actin is detected in the pellet fraction (P)). The adjustment of the pH to 8.0 facilitates F-actin depolymerization and the detection of monomeric actin in the supernatant (S). Cofilin preincubated with DMF in vitro (500 μm, equivalent to DMF:cofilin cysteines, 5:4) was unable to fully sever F-actin at pH 8.0 (Fig. 2C, 2D and 2E), resulting in a 29.3% decrease in actin severing activity). Supporting this, there was a greater proportion of actin (DMF: 53.49 ± 0.60% versus CONT: 34.20 ± 2.12%, n = 4, p < 0.01) and cofilin (DMF: 52.94 ± 0.84% versus CONT: 27.23 ± 3.25%, n = 4, p < 0.01) present in the pellet fraction (P) when the DMF modified cofilin was used (Fig. 2C, 2D and 2E). The specific modification of Cys139 in these in vitro cofilin preparations was also confirmed separately following CID (data available via PRIDE repository). These data suggest that the chemical modification of Cys139 on cofilin decreases its ability to dynamically regulate actin depolymerization.
Fig. 2.
Modification of cofilin impairs the depolymerization of actin filaments. A, MS/MS spectrum from astrocyte protein extracts after DMF treatment showing the MMF succinated Cys139 of cofilin 1 in the peptide HELQANCMMFYEEVKD. B, Confirmation that the cofilin severing effect on F-actin is pH-dependent. Samples of cofilin (7.5 μg, lanes 3–6) were incubated with F-actin (15 μg) in a buffer at pH 6.8 (lanes 3–4) or 8.0 (lanes 5–6); tubes containing actin only at pH 8.0 (lanes 1–2) were included as negative controls. After centrifugation, SDS/PAGE separation and Coomassie blue staining, the distribution of actin and cofilin was studied in the pellet after centrifugation (P) and the supernatant (S). Only the mix incubated at pH 8.0 showed cofilin severing activity on F-actin (lanes 5–6, note that the distribution of actin and cofilin is similar in P and S fractions); cofilin was inactive at pH 6.8 (lanes 3–4). C, Succination by DMF reduces cofilin severing effect on F-actin. Samples of cofilin (7.5 μg) were incubated with vehicle (0, lanes 1–8) or 500 μm DMF (500, lanes 9–16) and added to tubes containing F-actin (15 μg, lanes 1–17; lane 17 contained only actin as a negative control), and samples were processed as in (B). D and E, Quantification of the bands in (C), showing that exposure of cofilin to DMF decreases cofilin severing activity on F-actin. Results were expressed as mean ± S.E., with n = 4, ** p < 0.01 versus control (no DMF) by unpaired Student's t test.
Modification of Tubulin by DMF Does Not Alter Axonal Trafficking of Lysosomes
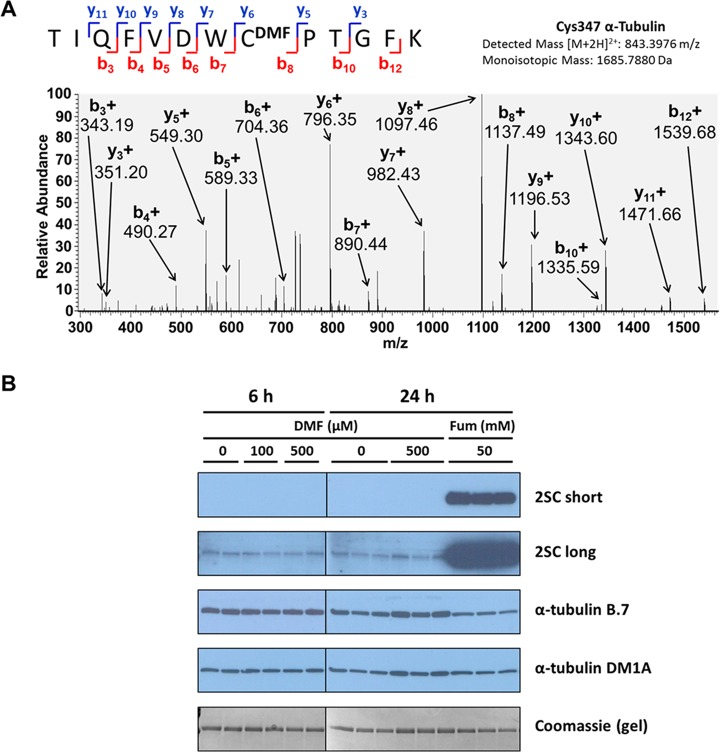
Tubulin was also identified as another cytoskeletal target of DMF reactivity (Table I). Tubulin α and β subunits form dimers that polymerize into microtubules, and we have previously described that in vitro treatment of purified porcine tubulin with DMF results in the succination of 11 of the 20 cysteines in the αβ tubulin dimer (30). In the current study we confirmed the DMF mediated succination of tubulin α-1A peptide TIQFVDWCDMFPTGFK (+2 charge state, 843.3976 m/z) in N1E-115 neurons; the site of succination was identified as Cys347 as designated by the prominent y6 and b8 fragment ions in the annotated spectrum (Fig. 3A). In addition, the pyridylethylated Cys347 containing peptide was also detected in the same N1E-115 neuronal protein preparation (+2 charge state, 823.9054 m/z, supplemental Fig. S3). We have previously described that increased succination by fumarate can affect the detectability of tubulin by antibodies directed against a cysteine containing antigen (30), as succination alters the epitope size and conveys two novel carboxylate groups. We have observed this in adipocytes cultured in high glucose (30 mm) versus normal glucose (5 mm), and on succinated porcine tubulin prepared in vitro (30). We examined if succination of purified porcine tubulin in vitro by DMF (versus fumarate) would also interfere with tubulin detection. Porcine tubulin was incubated with either 100 or 500 μm DMF, 500 mm fumarate, or without any addition for up to 24 h. Fig. 3B demonstrates that tubulin succination is pronounced in fumarate treated samples after a short film exposure (2SC short panel), whereas it is undetectable for the tubulin samples treated with DMF or left untreated. A longer exposure (Fig. 3B, 2SC long panel) shows some basal tubulin succination in untreated samples and a very strong signal in fumarate treated samples. However, we found no increases in modification of DMF treated tubulin compared with the untreated samples, which confirms the inability of the anti-2SC antibody to detect succination in the absence of ester hydrolysis, as shown above (Fig. 1A). After stripping the membranes, we probed the same blots with the α-tubulin antibody B-7, which is sensitive to tubulin succination (30). As shown in Fig. 3B (panel α-tubulin B-7), only the tubulin samples treated with fumarate showed a decrease in tubulin detectability, and this correlates with the increased succination detected by our 2SC antibody (Fig. 3B, panels 2SC short and long). Interestingly, tubulin samples treated with up to 500 μm DMF, though succinated (Fig. 3A and Table I), did not show decreased detectability by the α-tubulin antibody B-7 compared with controls (panel α-tubulin B-7), suggesting that succination by DMF, which leads to an uncharged modification of the cysteine residues, does not affect the interaction of succinated tubulin with the antibody. Moreover, pronounced succination of tubulin by either fumarate or DMF did not affect its detectability by the α-tubulin antibody DM1A, as already described, and we routinely use and recommend this antibody to detect total levels of tubulin in samples where succination may be present (30). Considering the abundance of commercially available antibodies against diverse epitopes for the same protein, it is important to examine the antigens used in order to establish reliable immunoblotting methods for total protein detection when post-translational modifications are endogenously present or introduced.
Fig. 3.
Tubulin as a target of DMF succination. A, MS/MS spectrum showing the DMF succinated Cys347 of α-tubulin in the peptide TIQFVDWCDMFPTGFK from N1E-115 neuron protein digests. B, Tubulin modification by DMF does not affect detectability by tubulin antibodies. 1 μg purified porcine brain tubulin was incubated for 6 or 24 h with 0 (control) or 100 or 500 μm DMF, or for 24 h with 50 mm fumarate (Fum) before SDS/PAGE separation and immunoblotting. If no saponification is performed, the anti-2SC antibody only detects succination by fumarate but not by DMF, even after a long exposure (2SC short and long panels; compare with Fig. 1). The α-tubulin B.7 antibody shows decreased detectability of tubulin after succination by fumarate, but no change when tubulin is modified by DMF, whereas the DM1A antibody is insensitive to succination (α-tubulin B.7 and DM1A panels, respectively). Coomassie staining of a duplicate gel run in parallel was used to verify even loading of the lanes.
Although the succination of tubulin by DMF did not affect tubulin interaction with the tested antibodies, we were interested in determining if tubulin interactions with motor proteins such as dynein or kinesin were altered, potentially impacting microtubule dynamics. Primary cultures of rat dorsal root ganglion (DRG) neurons were prepared and were treated with 50 μm DMF for 24 h or left untreated, and axonal lysosomal trafficking was monitored following lysosomal labeling with LysoTracker. Fig. 4A shows representative kymographs for the untreated DRG neurons (control panel) and DRG neurons treated with 50 μm DMF (50 μm DMF panel). No differences because of DMF treatment were observed in the percentage of lysosomes undergoing either anterograde or retrograde trafficking, or in the percentage of lysosomes changing directions (both), or of static lysosomes per axon (Fig. 4B). The average speed and run length of the motile lysosomes were also unchanged by DMF treatment (Fig. 4C). However, when motile events were separated according to different speeds (SP), DMF treatment changed the distribution of speed for retrograde events (Fig. 4D). Compared with control, there was higher percentage of 1–2 μm/sec motile events but lower percentage of 0.5–1 μm/sec motile events in DMF treated DRG neurons, with no changes in the run length (RL). The anterograde distribution of speed and run length were unaffected by DMF treatment. Overall the results indicate that lysosomal trafficking was not significantly altered, suggesting that DMF does not adversely affect this aspect of microtubule dynamics.
Collapsin Response Mediator 2 (CRMP2) as a Target of DMF Modification
All three cell preparations confirmed succination of the Collapsin Response Mediator 2 (CRMP2) peptide GLYDGPVCEVSVTPK by either MMF or DMF (Table I). The y8 and b8 fragment ion designations in the annotated spectrum shown in Fig. 5 confirm the designation of Cys504 as the site of succination by DMF in primary rat neurons (+2 charge state, 854.4109 m/z). The presence of pyridylethylated Cys504, representative of the unmodified peptide, was also detected in the primary rat neurons (+2 charge state, 834.9182 m/z, supplemental Fig. S4A). In addition, the MMF modification of Cys504 was also confirmed as shown by the annotated spectra of the GLYDGPVCMMFEVSVTPK peptide from DMF-treated primary astrocytes and DMF-treated N1E-115 cells (Fig. 5B and supplemental Fig. S4B, respectively). The C terminus of CRMP2 contains several GSK3β phosphorylation sites (Thr509, Thr514 and Ser 522) that have a role in mediating axonal growth cone retraction. The proximity of Cys504 to these phosphorylation sites, as well as the dependence of phosphorylation on Cys504 oxidation (40), suggest that site-specific succination of CRMP2 may contribute to DMF mediated axonal preservation (41).
Fig. 5.
CRMP-2 as a target of DMF modification in neural cells. A, MS/MS spectrum from rat primary neuron extracts after DMF treatment showing the DMF modified Cys504 of CRMP-2 in the peptide GLYDGPVCDMFEVSVTPK. B, MS/MS spectrum from rat primary astrocyte extracts after DMF treatment showing the MMF modified Cys504 of CRMP-2 in the peptide GLYDGPVCMMFEVSVTPK.
Ubiquitin C-Terminal Hydrolase L1 (Uchl1) Activity is Reduced
Ubiquitin C-Terminal hydrolase L1 (Uchl1) is an abundant deubiquitinase that constitutes up to 5% of total neuronal protein (42). Uchl1 cleaves ubiquitin from small peptide substrates and contributes to the regeneration of the intracellular ubiquitin pool (43). The modification of Uchl1 by both DMF and MMF was confirmed by a prominent y2 ion on the Cys152-containing peptide NEAIQAAHDSVAQEGQCR supplemental Fig. S5A and S5B), and this site has previously been shown to subject to chemical modification by other agents (44–46). DMF treatment did not alter the intracellular protein abundance of Uchl1 (detected using an antibody that specifically recognizes an N-terminal Uchl1 portion that did not contain any cysteines), nor did it alter the total levels of ubiquitinated protein by immunological detection supplemental Fig. S5C and S5E). UchL1 deubiquitinase activity was reduced in the presence of increasing concentrations of DMF (supplemental Fig. S5D, n = 5, p < 0.002 for 100 μm versus control), suggesting that further analyses are warranted to determine if the intracellular pools of free ubiquitin or Uchl1 structure are negatively impacted following exposure to DMF.
Mitochondrial Respiration is Minimally Altered by DMF
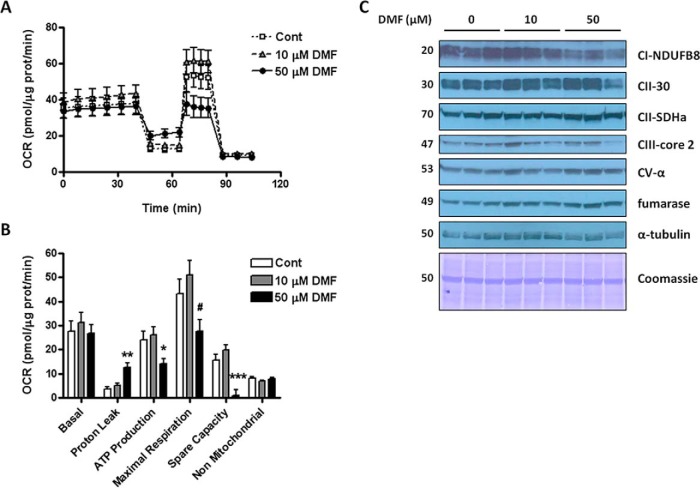
Because DMF treatment also resulted in succination of the abundant mitochondrial outer membrane protein voltage dependent anion channel-1 (VDAC-1, Table I), we used a Seahorse XF24 analyzer to measure the oxygen consumption rate (OCR) of N1E-115 neurons after treatment with 0–50 μm DMF for 24 h. Basal respiration was unaffected by DMF treatment at 10 and 50 μm, with 10 μm showing a trend toward slightly increased parameters of ATP synthesis, maximal respiration and the spare capacity. 50 μm DMF treatment resulted in decreases in the OCR linked to ATP synthesis, maximal respiration and the spare capacity, (Fig. 6A and 6B, this decrease was only statistically significant compared with the 10 μm DMF treatment rather than the control). Proton leak was increased only with 50 μm DMF (Fig. 6A and 6B). To further examine the decreased respiration because of 50 μm DMF, we also studied the levels of several components of the mitochondrial energetic machinery in N1E-115 neurons after treatment with 0–50 μm DMF for 24 h. As shown in Fig. 6C, only the expression of the electron transport chain (ETC) complex I subunit Ndufb8 was decreased by 50 μm DMF, whereas several other proteins related to mitochondrial energy production, including ETC complex II 30 kDa subunit and succinate dehydrogenase a, complex III core 2 subunit, complex V subunit α and tricarboxylic acid cycle fumarase, were unaffected by the 50 μm DMF treatment. Taken together, these results suggest that low dose 10 μm treatment with DMF does not affect the energy production machinery or respiration, whereas higher concentrations of DMF (50 μm) may lead to a partial decrease in some respiration parameters.
Fig. 6.
A and B, Respiratory profile of differentiated N1E-115 neurons after treatment with vehicle (open squares or open bars) or 10 μm (gray triangles or gray bars) or 50 μm (black circles or black bars) DMF for 24 h. The oxygen consumption rate (OCR) was measured and basal respiration (Basal), proton leak, oxygen consumption coupled to ATP production (ATP Production), maximal respiration, spare respiratory capacity (Spare Capacity), and nonmitochondrial respiration were calculated following the addition of medium, oligomycin, FCCP, and rotenone/antimycin A as described under Experimental Procedures. Data are representative of n = 6 measurements expressed as means ± S.E., ** p < 0.01 for 50 μm DMF versus Cont and 10 μm DMF; # p < 0.05 for 50 μm DMF versus 10 μm DMF (one way ANOVA and Tukey's post-test). C, Protein levels of select electron transport chain (ETC) subunits in N1E-115 neuronal cells after treatment with DMF. Total cell lysates (30 μg protein) from untreated N1E-115 cells (0) or cells treated with 10 or 50 μm DMF were separated by SDS/PAGE, transferred to a PVDF membrane and blotted with antibodies to the following ETC components: Complex I NDUFB8, Complex II subunit 30 and succinate dehydrogenase a (SDHa), Complex III core 2 subunit, and Complex V α-subunit. DMF treatment did not significantly change the levels of the ETC markers, except for CI-NDUFB8, which was decreased with 50 μm DMF. Fumarase and α-tubulin antibodies, as well as Coomassie staining of the membrane, were used to verify even loading, n = 3 per treatment group. The molecular masses of the proteins are indicated on the left-hand side.
DISCUSSION
Dimethyl fumarate (DMF) is an approved anti-inflammatory agent for the treatment of relapsing remitting multiple sclerosis and psoriasis. As an electrophile, DMF has been demonstrated to chemically modify cysteine residues in Keap1 and induce Nrf2-driven antioxidant response element gene transcription. Because DMF has been shown to improve the survival of neurons and astrocytes both in vivo and in vitro, we examined novel targets of DMF thiol reactivity in these cells types in order to better explain why DMF also offers benefit in Nrf2-knockout mouse models of inflammatory disease (17). Using both primary neurons and astrocytes, in addition to a neuronal cell line, we confirmed the identity of 27 uniquely modified cysteine residues, representing 24 distinct protein subunits in these cell types (Table I). We observed that DMF entered the cells and reacted rapidly with specific intracellular proteins, often with both methyl groups intact, indicating that DMF modifies many proteins before being hydrolyzed to the less reactive monomethyl fumarate (MMF) or fumarate itself. Fig. 1 further confirms that DMF is not completely hydrolyzed to fumarate inside the cell, as we could only detect succinated using an anti-2-succinocysteine antibody following base hydrolysis. Our results agree with the observations of Blewett et al., who recently described the reactivity of DMF versus MMF toward primary T cell proteins after 4 h of exposure to DMF in vitro (19). They indirectly quantified ∼40 DMF-sensitive cysteine residues, following detection of the cysteine residues that no longer reacted with the electrophile iodoacetamide-alkyne (isoTOP-ABPP method) (19) These residues were representative of ∼1% of the 2400 total cysteine residues identified, and confirm that fractional modification of functionally significant thiols is sufficient to contribute to altered T-cell activity (measured by reduced IL-2 production). In contrast to the isoTOP-ABPP, we searched directly for cysteine residues that had been variably modified by DMF, MMF or fumarate, in addition to pyridylethylated cysteines, and found that both DMF and MMF modified cysteine residues were detected across 24 proteins. Because our DMF incubation was for 24 h, it is possible that MMF may have more of an opportunity to be generated by esterase activity and then react with proteins. We did not detect any endogenously succinated residues directly modified by fumarate, however, because we were using positive ion mode mass spectrometry the detection of the negatively charged succinocysteine is more difficult to detect versus the fumarate esters. The longer exposure to DMF likely increased the probability of detecting modified proteins with extended half lives in the cell, such as the cytoskeletal protein subunits that were detected for tubulin, vimentin, glial fibrillary acid protein and cofilin-1. Exogenously applied DMF is more likely to react quickly with cytosolic proteins versus endogenously produced intramitochondrial fumarate that increases in other models such as fumarase deficient cancer cells (28). In the current study we observe the modification of only one mitochondrial protein by exogenous DMF, Voltage-Dependent Anion Channel-1, which is located on the outer mitochondrial membrane.
Cofilin-1 contributes to the remodeling of the cytoskeleton by depolymerizing filamentous (F) actin to provide actin monomers for growing actin filaments. We detected the modification of cofilin by MMF on Cys139 in astrocytes (Fig. 2A) and confirmed this site of modification when cofilin was incubated with DMF in vitro. The functional assessment of cofilin activity revealed that Cys139 modification resulted in a ∼30% decrease in cofilin's ability to depolymerize actin versus unmodified cofilin. DMF has been shown to enhance the differentiation of oligodendrocyte precursor cells (OPC) to oligodendrocytes, as evidenced by increased expression of O4 (47). Recently, Zuchero et al. described that early oligodendrocyte differentiation from OPCs is accompanied by the extension of processes containing ordered arrays of actin filaments, requiring polymerized actin and low cofilin activity (48). As the oligodendrocyte begins to mature further the actin cytoskeleton is disassembled, particularly by proteins such as cofilin, therefore differentiation requires dynamic changes in actin structure. Our data suggests that DMF-modified cofilin would be beneficial for actin arborization during early OPC differentiation. DMF may also act in a neuroprotective manner by inducing oligodendrocyte ensheathment of the axon, even if myelination is not dramatically increased. In addition, DMF treatment reduces localized microglial activation in models of neuroinflammation (49, 50), and cofilin has been shown to be critical for lipopolysaccharide mediated microglial activation as cofilin knockdown significantly inhibited microglial activity (51). Recently, magnetization transfer ratio measurements in humans have suggested that myelin density may be increased in those receiving delayed-release DMF treatment versus placebo (52), suggesting that DMF may beneficially modulate aspects of myelination during MS treatment. Together with the current literature, our data suggests that the DMF-mediated regulation of cofilin's actin-severing activity in dynamic glial cell populations, including oligodendrocytes, warrants further investigation.
Collapsin Response Mediator Protein 2 (CMRP2) is an intracellular phosphoprotein that contributes to axonal growth by transporting αβ tubulin heterodimers to the plus ends of the growing microtubule. The phosphorylation of CRMP2 leads to the collapse of the growth cone in response to environmental cues such as Semaphorin3A (Sema3A). CRMP2 is phosphorylated by Cdk5 at Ser522, and subsequently by GSK3β at Thr509, Thr514 and Ser518, leading to the disruption of the CRMP2:tubulin association and neurite retraction (53). In the current study, DMF contributed to the modification of Cys504 GLYDGPVCEVSVTPK in both primary neurons and N1E-115 derived neurons (Fig. 5A and supplemental Fig. S4A). Interestingly, the oxidation of Cys504 has been demonstrated to be critical for the Sema3A mediated recruitment of GSK3β (40). Sema3A signaling stimulates the production of intracellular hydrogen peroxide, leading to the oxidation and dimerization of adjacent Cys504. Thioredoxin (TRX) interacts to reduce the oxidized proteins and this CRMP2/TRX complex recruits GSK3β, resulting in CRMP2 phosphorylation and growth cone collapse (40). Because the oxidation of CRMP2 is necessary for GSK3β recruitment, it is possible that modification of this cysteine prevents oxidation-mediated phosphorylation. Consequently DMF treatment might be expected to preserve axonal integrity and prevent retraction in response to local degenerative cues. Enhanced phosphorylation of another CRMP2 site, Thr555 is abundant in active multiple sclerosis lesions from human autopsy (54). It is also significant that DMF, in combination with interferon β therapy, resulted in significant axonal preservation in a murine MS model (41). Our data suggests that CRMP2 Cys504 succination could be a significant mediator of the beneficial effects of DMF directly in neuronal cells, making it an attractive site for targeted therapies to prevent axonal loss.
We observed a trend toward the stimulation of mitochondrial respiration parameters following 10 μm DMF treatment of N1E neurons for 24 h, indicating that this dose does not acutely impair mitochondrial function. In contrast, 50 μm DMF tended to reduce ATP production, maximal respiration and spare respiratory capacity, however, this did not appear to be because of a loss of mitochondrial content as components of the electron transport chain (ETC) and tricarboxylic acid cycle examined were not affected (Fig. 6C). The exception to this was the decreased protein levels of the NDUFB8 subunit of Complex I, a 22kDa accessory protein that is required for the complete assembly of Complex I, it is possible that a loss of efficient electron transfer because of impaired Complex I assembly decreased NADH oxidation at this site. This may also explain why basal respiration appears to be unaffected by 50 μm DMF (Fig. 6A and 6B); FADH2 may sustain respiration under basal conditions, but the subsequent chemical challenges to determine maximal respiration and spare capacity put pressure on the ETC to increase electron transfer in cells where Complex I function may be compromised. The decrease in mitochondrial respiration observed with 50 μm DMF is like the decreased OCR parameters reported by Ahuja et al. using 20 μm DMF in murine embryonic fibroblasts for 24 h (55). Interestingly, Hayashi et al. have recently reported that 10 and 30 μm DMF increase mtDNA and maximal respiration after 48 h in human fibroblasts, and have shown tissue specific increases in mtDNA content (including the cerebellum) following 2 weeks of DMF administration to healthy mice (56). Taken together, the data suggests that variable doses and incubation times can have different effects on the models studied, although prolonged exposures in humans need to be monitored (57). In the current study we observed only a mild effect of DMF to alter lysosomal trafficking along microtubules in neurons, however in patients DMF may have more profound effects on other aspects of microtubule dynamics in cells directly exposed to the oral dosage.
In summary, although DMF is a prominent activator of Nrf2 mediated transcription, it also ameliorates experimental autoimmune encephalitis and the inflammatory status in Nrf2-knockout models. The novel discovery in this study that DMF and MMF may modify regulatory thiols on proteins such as cofilin-1 and CRMP2 suggest that DMF treatment may directly contribute to axonal preservation and remyelination, which is distinct from other MS treatments that are only known to modulate the inflammatory cell profile. Further investigation of the effects of DMF therapy on these novel neuroprotective targets in vivo will guide the development of more specific compounds for MS therapy.
DATA AVAILABILITY
The proteomic data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, http://www.ebi.ac.uk/pride, with the data set identifier PXD008314.
Supplementary Material
Acknowledgments
We thank Dr. Boris Kantor, USC Viral Vector Core for production of the lentiviral vectors and Professor John Baynes, University of South Carolina, for helpful discussion.
Footnotes
* This work was supported by the University of South Carolina Research Foundation ASPIRE-I award and the National Institutes of Health (R01 NS092938, R03 HD077187, R56 DK105087, P20 GM109091, F31 DK108559).
 This article contains supplemental Figures.
This article contains supplemental Figures.
1 The abbreviations used are:
- 2SC
- S-(2-succino)cysteine
- ARE
- antioxidant response element
- CID
- collision-induced dissociation
- CPE
- cysteine pyridylethylation
- C2SC
- cysteine succination by fumarate
- CMMF
- cysteine succination by monomethyl fumarate
- CDMF
- cysteine succination by dimethyl fumarate
- Ctx
- cortex
- CRMP2
- collapsin response mediator protein 2
- DC
- dendritic cells
- DDA
- data dependent acquisition
- DMF
- dimethyl fumarate
- DRG
- dorsal root ganglion
- GC
- gas chromatography
- GCL
- glutamate cysteine ligase
- GFAP
- glial fibrillary acid protein
- HCAR2
- hydroxycarboxylic acid receptor 2
- HLRCC
- hereditary leiomyomatosis and renal cell carcinoma
- HNE
- 4-hydroxy-2-nonenal
- HO1
- heme oxygenase 1
- IKKβ
- inhibitor of κβ kinase
- Keap1
- Kelch-like ECH-associated protein 1
- MOX
- methionine oxidation
- MMF
- monomethyl fumarate
- MRM
- multiple reaction monitoring
- MS
- multiple sclerosis
- NQO1
- NADPH-quinone-oxidoreductase-1
- Nrf2
- nuclear factor (erythroid-derived 2)-related factor 2
- OCR
- oxygen consumption rate
- OXPHOS
- oxidative phosphorylation
- PVDF
- polyvinylidene difluoride
- POX
- proline oxidation
- RIPA
- radioimmuno precipitation assay
- RRMS
- relapsing remitting multiple sclerosis
- SN
- sciatic nerve
- Str
- striatum
- VDAC
- voltage-dependent anion channel
- TNFAIP3
- tumor necrosis factor-α-induced protein 3.
REFERENCES
- 1. Mrowietz U., Christophers E., and Altmeyer P. (1998) Treatment of psoriasis with fumaric acid esters: results of a prospective multicentre study. German Multicentre Study. Br. J. Dermatol. 138, 456–460 [DOI] [PubMed] [Google Scholar]
- 2. Sheremata W., Brown A. D., and Rammohan K. W. (2015) Dimethyl fumarate for treating relapsing multiple sclerosis. Expert Opin. Drug Saf. 14, 161–170 [DOI] [PubMed] [Google Scholar]
- 3. Havrdova E., Giovannoni G., Gold R., Fox R. J., Kappos L., Phillips J. T., Okwuokenye M., and Marantz J. L. (2017) Effect of delayed-release dimethyl fumarate on no evidence of disease activity in relapsing-remitting multiple sclerosis: integrated analysis of the phase III DEFINE and CONFIRM studies. Eur. J. Neurol. 24, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee D. H., Stangel M., Gold R., and Linker R. A. (2013) The fumaric acid ester BG-12: a new option in MS therapy. Expert Rev. Neurother. 13, 951–958 [DOI] [PubMed] [Google Scholar]
- 5. Kappos L., Gold R., Arnold D. L., Bar-Or A., Giovannoni G., Selmaj K., Sarda S. P., Agarwal S., Zhang A., Sheikh S. I., Seidman E., and Dawson K. T. (2014) Quality of life outcomes with BG-12 (dimethyl fumarate) in patients with relapsing-remitting multiple sclerosis: the DEFINE study. Mult. Scler. 20, 243–252 [DOI] [PubMed] [Google Scholar]
- 6. Treumer F., Zhu K., Glaser R., and Mrowietz U. (2003) Dimethylfumarate is a potent inducer of apoptosis in human T cells. J. Invest. Dermatol. 121, 1383–1388 [DOI] [PubMed] [Google Scholar]
- 7. Hoxtermann S., Nuchel C., and Altmeyer P. (1998) Fumaric acid esters suppress peripheral CD4- and CD8-positive lymphocytes in psoriasis. Dermatology 196, 223–230 [DOI] [PubMed] [Google Scholar]
- 8. de Jong R., Bezemer A. C., Zomerdijk T. P., van de Pouw-Kraan T., Ottenhoff T. H., and Nibbering P. H. (1996) Selective stimulation of T helper 2 cytokine responses by the anti-psoriasis agent monomethylfumarate. Eur. J. Immunol. 26, 2067–2074 [DOI] [PubMed] [Google Scholar]
- 9. Ghoreschi K., Bruck J., Kellerer C., Deng C., Peng H., Rothfuss O., Hussain R. Z., Gocke A. R., Respa A., Glocova I., Valtcheva N., Alexander E., Feil S., Feil R., Schulze-Osthoff K., Rupec R. A., Lovett-Racke A. E., Dringen R., Racke M. K., and Rocken M. (2011) Fumarates improve psoriasis and multiple sclerosis by inducing type II dendritic cells. J. Exp. Med. 208, 2291–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt M. M., and Dringen R. (2010) Fumaric acid diesters deprive cultured primary astrocytes rapidly of glutathione. Neurochem. Int. 57, 460–467 [DOI] [PubMed] [Google Scholar]
- 11. Lehmann J. C., Listopad J. J., Rentzsch C. U., Igney F. H., von Bonin A., Hennekes H. H., Asadullah K., and Docke W. D. (2007) Dimethylfumarate induces immunosuppression via glutathione depletion and subsequent induction of heme oxygenase 1. J. Invest. Dermatol. 127, 835–845 [DOI] [PubMed] [Google Scholar]
- 12. Thiessen A., Schmidt M. M., and Dringen R. (2010) Fumaric acid dialkyl esters deprive cultured rat oligodendroglial cells of glutathione and upregulate the expression of heme oxygenase 1. Neurosci. Lett. 475, 56–60 [DOI] [PubMed] [Google Scholar]
- 13. Linker R. A., Lee D. H., Ryan S., van Dam A. M., Conrad R., Bista P., Zeng W., Hronowsky X., Buko A., Chollate S., Ellrichmann G., Bruck W., Dawson K., Goelz S., Wiese S., Scannevin R. H., Lukashev M., and Gold R. (2011) Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain 134, 678–692 [DOI] [PubMed] [Google Scholar]
- 14. Gopal S., Mikulskis A., Gold R., Fox R. J., Dawson K. T., and Amaravadi L. (2017) Evidence of activation of the Nrf2 pathway in multiple sclerosis patients treated with delayed-release dimethyl fumarate in the Phase 3 DEFINE and CONFIRM studies. Mult. Scler 1352458517690617 [DOI] [PubMed] [Google Scholar]
- 15. Bruck W., Gold R., Lund B. T., Oreja-Guevara C., Prat A., Spencer C. M., Steinman L., Tintore M., Vollmer T. L., Weber M. S., Weiner L. P., Ziemssen T., and Zamvil S. S. (2013) Therapeutic decisions in multiple sclerosis: moving beyond efficacy. JAMA Neurol. 70, 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fox R. J., Kita M., Cohan S. L., Henson L. J., Zambrano J., Scannevin R. H., O'Gorman J., Novas M., Dawson K. T., and Phillips J. T. (2014) BG-12 (dimethyl fumarate): a review of mechanism of action, efficacy, and safety. Curr. Med. Res. Opin. 30, 251–262 [DOI] [PubMed] [Google Scholar]
- 17. Schulze-Topphoff U., Varrin-Doyer M., Pekarek K., Spencer C. M., Shetty A., Sagan S. A., Cree B. A., Sobel R. A., Wipke B. T., Steinman L., Scannevin R. H., and Zamvil S. S. (2016) Dimethyl fumarate treatment induces adaptive and innate immune modulation independent of Nrf2. Proc. Natl. Acad. Sci. U.S.A. 113, 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen H., Assmann J. C., Krenz A., Rahman M., Grimm M., Karsten C. M., Kohl J., Offermanns S., Wettschureck N., and Schwaninger M. (2014) Hydroxycarboxylic acid receptor 2 mediates dimethyl fumarate's protective effect in EAE. J. Clin. Invest. 124, 2188–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blewett M. M., Xie J., Zaro B. W., Backus K. M., Altman A., Teijaro J. R., and Cravatt B. F. (2016) Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci. Signal. 9, rs10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Piroli G. G., Manuel A. M., Clapper A. C., Walla M. D., Baatz J. E., Palmiter R. D., Quintana A., and Frizzell N. (2016) Succination is increased on select proteins in the brainstem of the NADH dehydrogenase (ubiquinone) Fe-S protein 4 (Ndufs4) knockout mouse, a model of Leigh syndrome. Mol. Cell. Proteomics 15, 445–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alderson N. L., Wang Y., Blatnik M., Frizzell N., Walla M. D., Lyons T. J., Alt N., Carson J. A., Nagai R., Thorpe S. R., and Baynes J. W. (2006) S-(2-Succinyl)cysteine: a novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch. Biochem. Biophys. 450, 1–8 [DOI] [PubMed] [Google Scholar]
- 22. Nagai R., Brock J. W., Blatnik M., Baatz J. E., Bethard J., Walla M. D., Thorpe S. R., Baynes J. W., and Frizzell N. (2007) Succination of protein thiols during adipocyte maturation: a biomarker of mitochondrial stress. J. Biol. Chem. 282, 34219–34228 [DOI] [PubMed] [Google Scholar]
- 23. Frizzell N., Rajesh M., Jepson M. J., Nagai R., Carson J. A., Thorpe S. R., and Baynes J. W. (2009) Succination of thiol groups in adipose tissue proteins in diabetes: succination inhibits polymerization and secretion of adiponectin. J. Biol. Chem. 284, 25772–25781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas S. A., Storey K. B., Baynes J. W., and Frizzell N. (2012) Tissue distribution of S-(2-succino)cysteine (2SC), a biomarker of mitochondrial stress in obesity and diabetes. Obesity 20, 263–269 [DOI] [PubMed] [Google Scholar]
- 25. Merkley E. D., Metz T. O., Smith R. D., Baynes J. W., and Frizzell N. (2014) The succinated proteome. Mass Spectrom. Rev. 33, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bardella C., El-Bahrawy M., Frizzell N., Adam J., Ternette N., Hatipoglu E., Howarth K., O'Flaherty L., Roberts I., Turner G., Taylor J., Giaslakiotis K., Macaulay V. M., Harris A. L., Chandra A., Lehtonen H. J., Launonen V., Aaltonen L. A., Pugh C. W., Mihai R., Trudgian D., Kessler B., Baynes J. W., Ratcliffe P. J., Tomlinson I. P., and Pollard P. J. (2011) Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 225, 4–11 [DOI] [PubMed] [Google Scholar]
- 27. Chen Y. B., Brannon A. R., Toubaji A., Dudas M. E., Won H. H., Al-Ahmadie H. A., Fine S. W., Gopalan A., Frizzell N., Voss M. H., Russo P., Berger M. F., Tickoo S. K., and Reuter V. E. (2014) Hereditary leiomyomatosis and renal cell carcinoma syndrome-associated renal cancer: recognition of the syndrome by pathologic features and the utility of detecting aberrant succination by immunohistochemistry. Am. J. Surg. Pathol. 38, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adam J., Hatipoglu E., O'Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G. W., Wolhuter K., Stevens M., Fischer R., Carmeliet P., Maxwell P. H., Pugh C. W., Frizzell N., Soga T., Kessler B. M., El-Bahrawy M., Ratcliffe P. J., and Pollard P. J. (2011) Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20, 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manuel A. M., and Frizzell N. (2013) Adipocyte protein modification by Krebs cycle intermediates and fumarate ester-derived succination. Amino Acids 45, 1243–1247 [DOI] [PubMed] [Google Scholar]
- 30. Piroli G. G., Manuel A. M., Walla M. D., Jepson M. J., Brock J. W., Rajesh M. P., Tanis R. M., Cotham W. E., and Frizzell N. (2014) Identification of protein succination as a novel modification of tubulin. Biochem. J. 462, 231–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brennan M. S., Patel H., Allaire N., Thai A., Cullen P., Ryan S., Lukashev M., Bista P., Huang R., Rhodes K. J., and Scannevin R. H. (2016) Pharmacodynamics of dimethyl fumarate are tissue specific and involve NRF2-dependent and -independent mechanisms. Antioxid. Redox Signal. 24, 1058–1071 [DOI] [PubMed] [Google Scholar]
- 32. Brewer G. J., and Torricelli J. R. (2007) Isolation and culture of adult neurons and neurospheres. Nat. Protoc. 2, 1490–1498 [DOI] [PubMed] [Google Scholar]
- 33. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 34. Oh J. E., Karlmark K. R., Shin J. H., Pollak A., Freilinger A., Hengstschlager M., and Lubec G. (2005) Differentiation of neuroblastoma cell line N1E-115 involves several signaling cascades. Neurochem. Res. 30, 333–348 [DOI] [PubMed] [Google Scholar]
- 35. Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pandey J. P., and Smith D. S. (2011) A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J. Neurosci. 31, 17207–17219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yonezawa N., Nishida E., and Sakai H. (1985) pH control of actin polymerization by cofilin. J. Biol. Chem. 260, 14410–14412 [PubMed] [Google Scholar]
- 38. Tanis R. M., Piroli G. G., Day S. D., and Frizzell N. (2015) The effect of glucose concentration and sodium phenylbutyrate treatment on mitochondrial bioenergetics and ER stress in 3T3-L1 adipocytes. Biochim. Biophys. Acta 1853, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manuel A. M., Walla M. D., Faccenda A., Martin S. L., Tanis R. M., Piroli G. G., Adam J., Kantor B., Mutus B., Townsend D. M., and Frizzell N. (2017) Succination of Protein Disulfide Isomerase Links Mitochondrial Stress and Endoplasmic Reticulum Stress in the Adipocyte During Diabetes. Antioxid. Redox Signal. 27, 1281–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morinaka A., Yamada M., Itofusa R., Funato Y., Yoshimura Y., Nakamura F., Yoshimura T., Kaibuchi K., Goshima Y., Hoshino M., Kamiguchi H., and Miki H. (2011) Thioredoxin mediates oxidation-dependent phosphorylation of CRMP2 and growth cone collapse. Sci Signal 4, ra26. [DOI] [PubMed] [Google Scholar]
- 41. Reick C., Ellrichmann G., Thone J., Scannevin R. H., Saft C., Linker R. A., and Gold R. (2014) Neuroprotective dimethyl fumarate synergizes with immunomodulatory interferon beta to provide enhanced axon protection in autoimmune neuroinflammation. Exp. Neurol. 257, 50–56 [DOI] [PubMed] [Google Scholar]
- 42. Bishop P., Rocca D., and Henley J. M. (2016) Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem. J. 473, 2453–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bett J. S., Ritorto M. S., Ewan R., Jaffray E. G., Virdee S., Chin J. W., Knebel A., Kurz T., Trost M., Tatham M. H., and Hay R. T. (2015) Ubiquitin C-terminal hydrolases cleave isopeptide- and peptide-linked ubiquitin from structured proteins but do not edit ubiquitin homopolymers. Biochem. J. 466, 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koharudin L. M., Liu H., Di Maio R., Kodali R. B., Graham S. H., and Gronenborn A. M. (2010) Cyclopentenone prostaglandin-induced unfolding and aggregation of the Parkinson disease-associated UCH-L1. Proc. Natl. Acad. Sci. U.S.A. 107, 6835–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Toyama T., Abiko Y., Katayama Y., Kaji T., and Kumagai Y. (2015) S-Mercuration of ubiquitin carboxyl-terminal hydrolase L1 through Cys152 by methylmercury causes inhibition of its catalytic activity and reduction of monoubiquitin levels in SH-SY5Y cells. J. Toxicol. Sci. 40, 887–893 [DOI] [PubMed] [Google Scholar]
- 46. Toyama T., Shinkai Y., Yazawa A., Kakehashi H., Kaji T., and Kumagai Y. (2014) Glutathione-mediated reversibility of covalent modification of ubiquitin carboxyl-terminal hydrolase L1 by 1,2-naphthoquinone through Cys152, but not Lys4. Chem. Biol. Interact. 214, 41–48 [DOI] [PubMed] [Google Scholar]
- 47. Galloway D. A., Williams J. B., and Moore C. S. (2017) Effects of fumarates on inflammatory human astrocyte responses and oligodendrocyte differentiation. Ann. Clin. Transl. Neurol. 4, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zuchero J. B., Fu M. M., Sloan S. A., Ibrahim A., Olson A., Zaremba A., Dugas J. C., Wienbar S., Caprariello A. V., Kantor C., Leonoudakis D., Lariosa-Willingham K., Kronenberg G., Gertz K., Soderling S. H., Miller R. H., and Barres B. A. (2015) CNS myelin wrapping is driven by actin disassembly. Dev. Cell 34, 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peng H., Li H., Sheehy A., Cullen P., Allaire N., and Scannevin R. H. (2016) Dimethyl fumarate alters microglia phenotype and protects neurons against proinflammatory toxic microenvironments. J. Neuroimmunol. 299, 35–44 [DOI] [PubMed] [Google Scholar]
- 50. Fowler J. H., McQueen J., Holland P. R., Manso Y., Marangoni M., Scott F., Chisholm E., Scannevin R. H., Hardingham G. E., and Horsburgh K. (2017) Dimethyl fumarate improves white matter function following severe hypoperfusion: Involvement of microglia/macrophages and inflammatory mediators. J. Cereb. Blood Flow Metab. 271678x17713105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alhadidi Q., and Shah Z. A. (2017) Cofilin Mediates LPS-Induced Microglial Cell Activation and Associated Neurotoxicity Through Activation of NF-kappaB and JAK-STAT Pathway. Mol. Neurobiol. 55, 1676–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arnold D. L., Gold R., Kappos L., Bar-Or A., Giovannoni G., Selmaj K., Yang M., Zhang R., Stephan M., Sheikh S. I., and Dawson K. T. (2014) Magnetization transfer ratio in the delayed-release dimethyl fumarate DEFINE study. J. Neurol. 261, 2429–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmidt E. F., and Strittmatter S. M. (2007) The CRMP family of proteins and their role in Sema3A signaling. Adv. Exp. Med. Biol. 600, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Petratos S., Ozturk E., Azari M. F., Kenny R., Lee J. Y., Magee K. A., Harvey A. R., McDonald C., Taghian K., Moussa L., Mun Aui P., Siatskas C., Litwak S., Fehlings M. G., Strittmatter S. M., and Bernard C. C. (2012) Limiting multiple sclerosis related axonopathy by blocking Nogo receptor and CRMP-2 phosphorylation. Brain 135, 1794–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ahuja M., Ammal Kaidery N., Yang L., Calingasan N., Smirnova N., Gaisin A., Gaisina I. N., Gazaryan I., Hushpulian D. M., Kaddour-Djebbar I., Bollag W. B., Morgan J. C., Ratan R. R., Starkov A. A., Beal M. F., and Thomas B. (2016) Distinct Nrf2 Signaling mechanisms of fumaric acid esters and their role in neuroprotection against 1-Methyl-4-Phenyl-1,2,3,6-tetrahydropyridine-induced experimental Parkinson's-like disease. J. Neurosci. 36, 6332–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hayashi G., Jasoliya M., Sacca F., Pane C., Filla A., Marsili A., Puorro G., Lanzillo R., Brescia Morra V., and Cortopassi G. (2017) Dimethyl fumarate mediates Nrf2-dependent mitochondrial biogenesis in mice and humans. Hum. Mol. Genet. 26, 2864–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haghikia A., Perrech M., Pula B., Ruhrmann S., Potthoff A., Brockmeyer N. H., Goelz S., Wiendl H., Linda H., Ziemssen T., Baranzini S. E., Kall T. B., Bengel D., Olsson T., Gold R., and Chan A. (2011) Functional energetics of CD4+-cellular immunity in monoclonal antibody-associated progressive multifocal leukoencephalopathy in autoimmune disorders. PLoS ONE 6, e18506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomic data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, http://www.ebi.ac.uk/pride, with the data set identifier PXD008314.