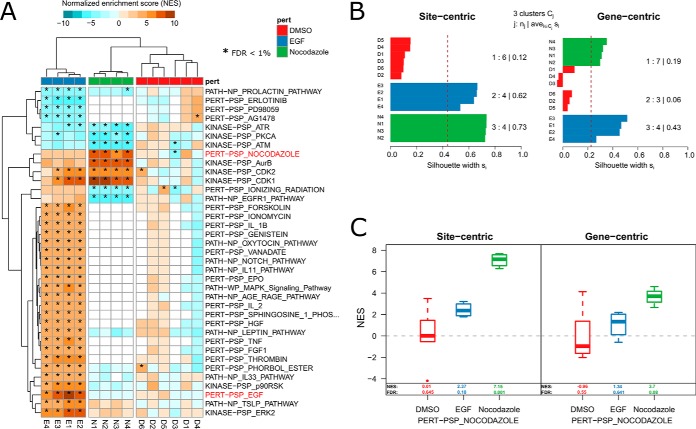
Fig. 3.
Phosphoproteome signatures of EGF and nocodazole treatment in HeLa cells. We applied PTM-SEA to a phosphoproteome data set from the literature. Sharma et al. (18) studied phosphoproteome dynamics of EGF treatment in HeLa S3 cells. To mitotically arrest the cells, the antineoplastic agent nocodazole was used. Because PTMsigDB contained signatures for both perturbations, this data set was used as a benchmark to evaluate PTM-SEA. A, Heatmap depicting normalized enrichment scores (NES) of signatures (rows) in PTMsigDB that were consistently enriched or depleted at FDR < 0.01 in replicate measurements of EGF/nocodazole treatments (marked by asterisks). Hierarchical clustering of enrichment scores separated samples (columns) by experimental condition (EGF, nocodazole and control). Enrichment scores of nocodazole and EGF signature showed highest magnitude in the respective experiments and are highlighted in red. B, Silhouette analysis comparing hierarchical clustering of site-centric (left panel) and gene-centric (right panel) signature enrichment scores into three clusters. The bar chart depicts silhouette scores (x axis) of each sample (y axis) colored according to the assigned cluster. Average silhouette scores for each cluster are depicted at the right side of the bar charts. C, Signature enrichment scores of the nocodazole perturbation signature calculated using site-centric (left panel) and gene-centric (right panel) approach. Numbers indicate the median NES and FDR in replicate measurements of respective perturbations (x axis).