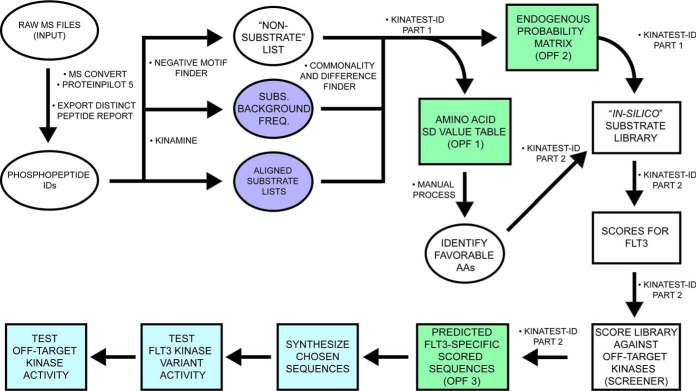
Fig. 2.
Conceptual overview of our KALIP data processing and formatting for KINATEST-ID incorporation to develop FLT3 artificial substrates. After conversion and peptide/protein ID in the Galaxy-P proteomic pipeline, the KinaMine data formatter tool extracted the confidently-identified (1% FDR) sequences that contained a phosphorylated tyrosine residue (pY), centrally aligned the sequences to the tyrosine of interest (“Aligned substrate lists”) and extracted the UniProt accession number with the accompanying proteins' amino acid composition file (“Subs. Background Freq.”). The Negative motif finder script uses the Uniprot accession numbers to generate a list of tyrosine-containing potential tryptic peptides that were not observed in the phosphopeptide data set. The scripts Kinatest-R part1.r and -part2.r processed those input files to identify substrate preferences and generate a ranked list of candidate sequences as potential FLT3 substrates. Kinatest-R part2.r then scores the input substrate and non-substrate lists and outputs as two additional files. After performing this workflow on phosphopeptide data from FLT3 kinase reactions, chosen candidate sequences were synthesized and validated in vitro against the FLT3 kinase variants. The candidate sequences were then assayed in vitro against a panel of kinases to determine off-target kinase activity. In silico/predictive steps are illustrated in white/green. Empirical steps of synthesizing peptides and characterizing FLT3 activity and specificity are depicted in light blue.