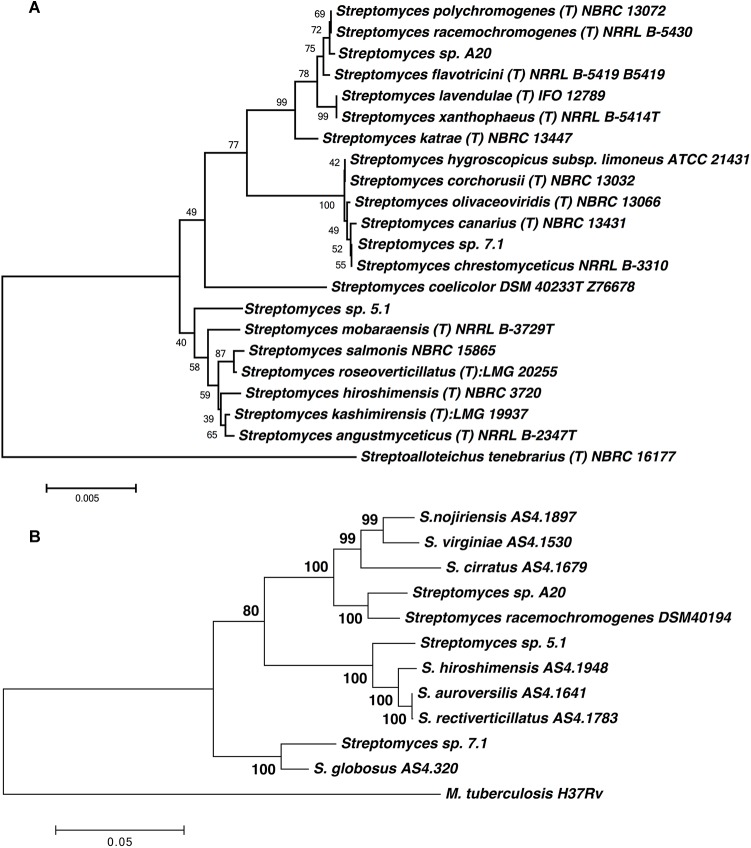
FIGURE 1.
(A) Phylogenetic tree based on 16S rRNA sequences of the Streptomyces strains. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Numbers at branch nodes are bootstrap values. Sequence from Streptoalloteichus tenebrarius NBRC 16177 was included as outgroup. (B) Phylogenetic tree based on MLST analyses from five loci (gyrB, atpD, recA trpB, and rpoB). The tree was drawn to scale using Maximum Likelihood method and Tamura-Nei model, with branch lengths measured in the number of substitutions per site. The analysis involved 12 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 2896 positions in the final dataset.