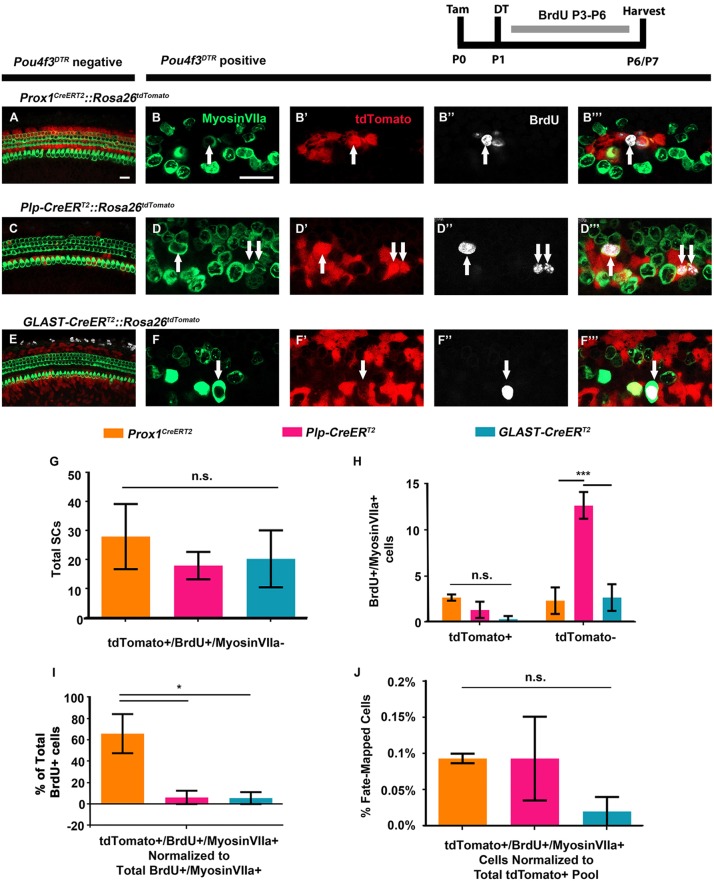
Fig. 7.
A larger proportion of mitotically regenerated HCs arise from PCs and DCs. (A-F‴) Representative confocal slice images from P6-P7 Prox1CreERT2::Rosa26tdTomato::Pou4f3DTR (B-B‴), Plp-CreERT2::Rosa26tdTomato::Pou4f3DTR (D-D‴) and GLAST-CreERT2::Rosa26tdTomato::Pou4f3DTR (F-F‴) mice, as well as littermate controls that lacked Pou4f3DTR (A,C,E). All mice were injected with tamoxifen (Tam) at P0 to induce tdTomato expression in various SC groups, with diphtheria toxin (DT) at P1 to induce HC death in the experimental samples, and with BrdU twice per day from P3-P6 (∼6 h between injections) to detect dividing cells. Samples were immunostained with anti-myosin VIIa antibodies to label HCs (green), anti-BrdU antibodies to label mitotic cells (white) and anti-RFP antibodies to detect tdTomato expression (red). White arrows indicate tdTomato-positive/BrdU-positive/myosin VIIa-positive mitotically regenerated, fate-mapped HCs. Scale bars: 25 μm. (G) No difference was observed in the number of BrdU-positive, fate-mapped SCs that remained as SCs (tdTomato-positive/BrdU-positive/myosin VIIa-negative) among CreER lineages. (H) Significantly more mitotically regenerated HCs that were not fate-mapped (BrdU-positive/myosinVIIA-positive/tdTomato-negative cells) were observed in Plp-CreERT2::Rosa26tdTomato::Pou4f3DTR mice compared with both Prox1CreERT2 and GLAST-CreERT2 lineages. However, there was no difference in the number of fate-mapped, mitotically regenerated HCs (BrdU-positive/myosin VIIA-positive/tdTomato-positive cells) among the three lines. (I) When normalized to the total number of BrdU-positive cells, a larger percentage of BrdU-positive/myosin VIIa-positive cells were fate-mapped (tdTomato-positive) by the Prox1CreERT2 line, compared with Plp-CreERT2 and GLAST-CreERT2 lineages. (J) When the total number of fate-mapped mitotically regenerated HCs was normalized to the total number of tdTomato-positive SCs in control cochleae for each CreER line, all groups were found to have an equal capacity for mitotic regeneration. *P<0.05, ***P<0.001, determined using two-way ANOVA with a Tukey's post-hoc test. Data are mean±s.e.m.; n=3. n.s., not significant.