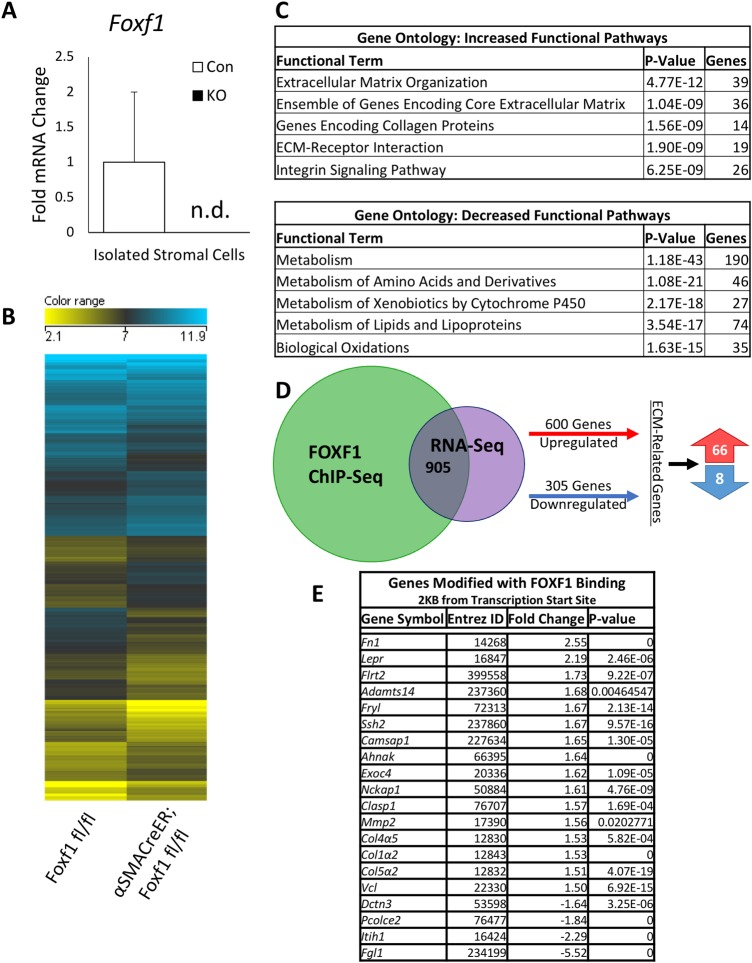
Fig. 5.
FOXF1 deletion alters expression of pro-fibrotic genes in hepatic myofibroblasts. (A) qRT-PCR analysis of primary hepatic stromal cells submitted for RNA sequencing shows that Foxf1 mRNA is not detected (n.d.) in αSMACreER;Foxf1−/− livers. Samples were pooled for further analysis. (B) Heat map shows differentially expressed genes in stromal cells from Foxf1fl/fl and αSMACreER;Foxf1−/− livers after chronic CCl4-induced hepatic injury as identified by RNA-seq analysis. (C) Biological processes influenced by the deletion of Foxf1 were identified using ToppFunn. P-values and number of genes are listed for each classification. (D) 905 overlapping genes were identified between ChIP-seq (GEO accession GSE100149) and RNA-seq (GEO accession GSE123726) data, of which 74 were ECM-related genes. (E) Table shows 20 ECM-related genes identified by ChIP-seq and RNA-seq (FOXF1 binding was analyzed within 2 KB from transcriptional start site).