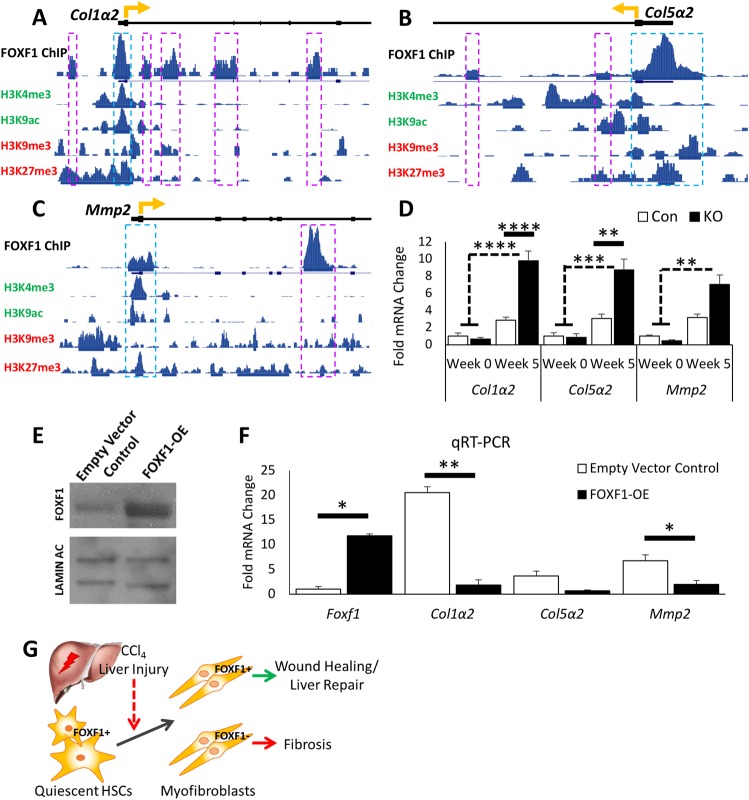
Fig. 6.
FOXF1 binds to DNA regulatory regions of Col1α2, Col5α2 and Mmp2. (A–C) ChIP-seq shows FOXF1 binding near the transcriptional start sites in Col1α2, Col5α2 and Mmp2 gene loci. Histone modification marks of enhancers (H3K4me3, H3K9ac) and repressors (H3K9me3, H3K27me3) are aligned with FOXF1-binding regions. Significant areas of FOXF1 binding are marked with boxes, with blue boxes indicating the binding site is within gene promoter region. Gene transcriptional start sites are marked with a directional yellow arrow. (D) qRT-PCR analysis shows the significant increase of Col1α2, Col5α2 and Mmp2 mRNAs in the isolated stromal cells of CCl4-treated αSMACreER;Foxf1−/− livers. For Col1α2 and Col5α2: n=3 mice per group in week 0; n=5 mice per group in week 5. For Mmp2: n=3 mice per group in week 0; n=6 control mice and n=4 KO mice in week 5. (E) Western blot shows increase in FOXF1 expression in isolated HSCs after FOXF1-overexpression. Cropped blots are presented here with full length blots presented in Fig. S12. (F) qRT-PCR shows an increase of Foxf1 mRNA and a decrease of Col1α2, Col5α2 and Mmp2 mRNAs in isolated HSCs after FOXF1-overexpression. (G) Diagram of hepatic fibrosis in Foxf1-deficient mice shows that the loss of FOXF1 promotes ECM deposition and exacerbated fibrosis after CCl4-treatment. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.