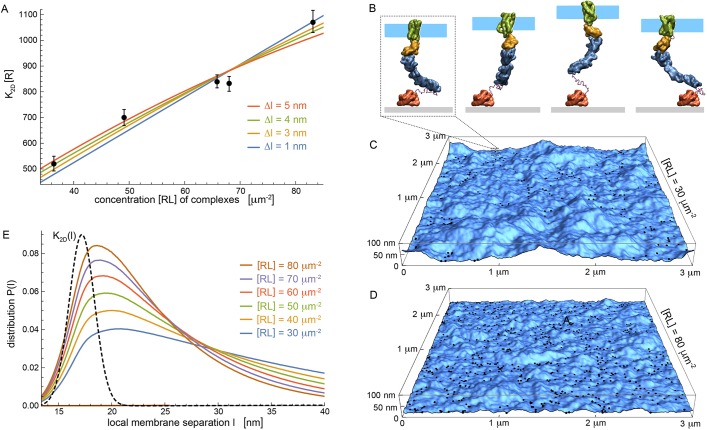
Fig. 2.
Multiscale modeling of experiments to clarify interactions. (A) Binding constant K2D as a function of complex concentration [RL] from experiments (data points) and modeling (lines) for different values Δl of repulsive membrane-substrate interactions. Modeling is based on Eqn 1, with fit values Kmax=(956±24)/[R], (470±10)/[R], (378±8)/[R] and (326±8)/[R] for Δl=1, 3, 4, 5 nm, respectively. Binding constant K2D has units of concentration [R] of unbound SIRPα. Error bars indicate 95% confidence intervals for fitting the reaction-diffusion model±s.e.m.). Each data point corresponds to measurement of an individual GPMV in at least two independent experiments. (B) Simulation snapshots of coarse-grained CD47-SIRPα complex. Separation between fluid membrane patch (cyan) and substrate (grey) varies in simulations mainly because of conformational changes of the unstructured linker that covalently connects extracelluar SIRPα domain (blue) to substrate-bound GST (red). (C,D) Snapshots of adhering membranes with area 3×3 μm2 for respective concentrations [RL]=30, 80 μm−2 of CD47-SIRPα complexes at Δl=4 nm. Black dots indicate complexes. (E) Distributions P(l) for local separation between membrane and substrate obtained from averaging over many membrane snapshots for various [RL]. Dashed black line represents K2D(l) (arbitrary units).