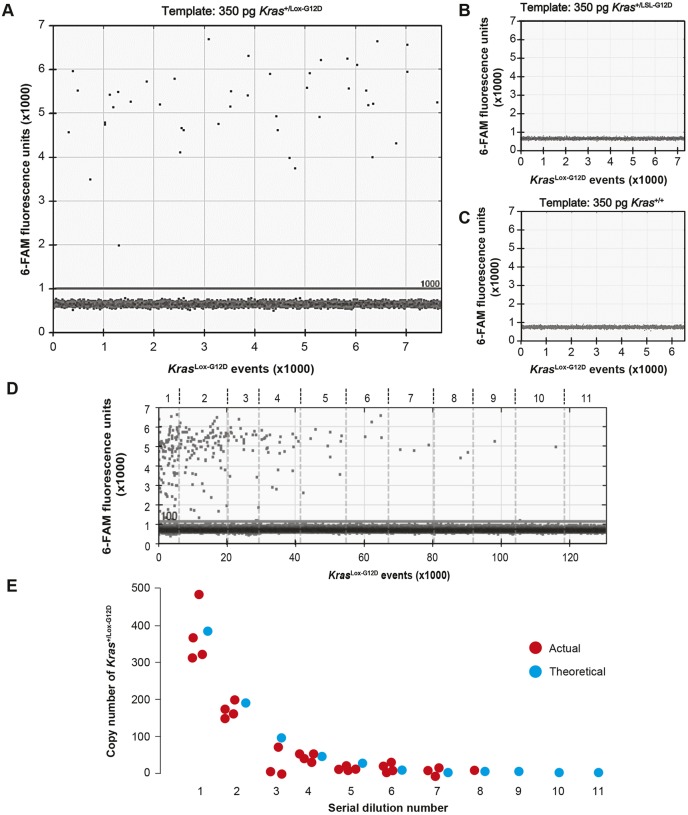
Fig. 4.
Validation of ddPCR assay for detection of KrasLox-G12D allele using LNA-2 primers. (A-C) One-dimensional droplet plot of LNA-2 assay (comprising probe LNA-2 and primers flanking the recombined LoxP sequence in KrasLox-G12D) with Kras+/Lox-G12D MEF genomic DNA (A), Kras+/LSL-G12D MEF genomic DNA (B) and Kras+/+ MEF genomic DNA (C). Primers were annealed at 62°C, following a gradient PCR experiment to determine the optimum annealing temperature (Ta, not shown). A manual threshold of 1000 fluorescence units was selected. (D) Kras+/Lox-G12D MEF genomic DNA was serially diluted into a background of Kras+/LSL-G12D MEF genomic DNA. A one-dimensional droplet plot for the 133-bp LNA-2 assay at each serial dilution is shown. A manual threshold of 1100 fluorescence units was selected. (E) Actual versus theoretical copy number of KrasLox-G12D at each serial dilution.