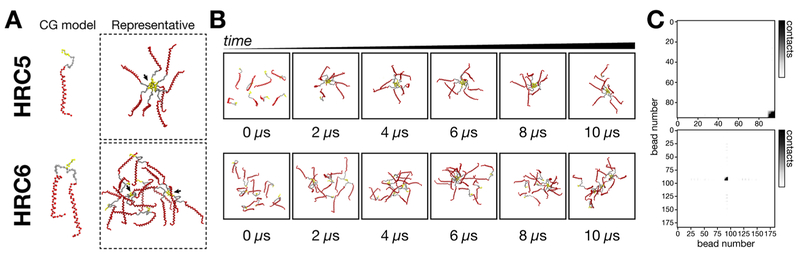
Figure 4.
CG MD simulations of HRC5 and HRC6 assembly into NP. Coarse-grained molecular dynamics simulations of 8 HRC5 or HRC6 peptides, followed for 10 μs. (A) Representative image of the peptide models and most characteristic peptide assembling behavior. The peptide chain backbone is represented in red, the PEG4 linker in gray, and the Toc moiety in yellow (all other system components were omitted for clarity). Black arrows indicate different association clusters. (B) Stills of each peptide simulation, captured at 0, 2, 4, 6, 8, and 10 μs, collected to emphasize the progression of the peptides assembly during the experiment. (C) Preferential contact maps reporting favored interaction regions between peptides, built from full simulation data. For HRC5, the CG bead numbers correspond to peptide chain −0 to 80; PEG4 linker −81 to 89; and Toc −90 to 95. For HRC6, CG bead numbers are the same as those for HRC5, with the second peptide chain and PEG4 linker corresponding to beads 96 through 180. Darker tones in the contact map gradient scale indicate a higher number of detected contacts. For both HRC5 and HRC6, Toc−Toc interactions can be seen to predominate; additional Toc−peptide interactions are visible for HRC6 but not for HRC5.