FIGURE 3.
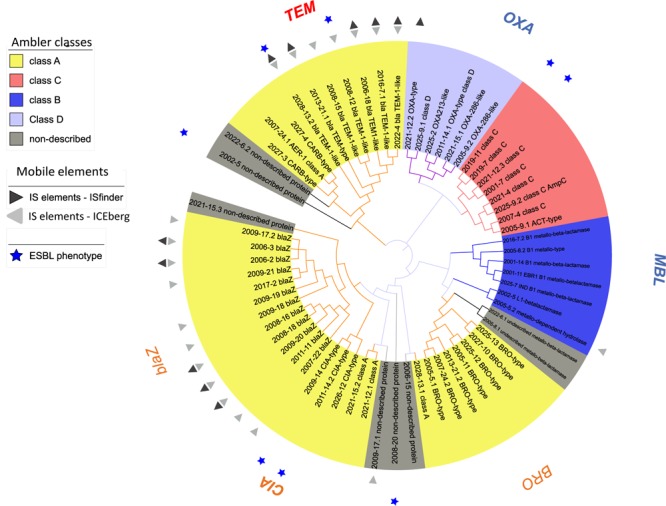
Phylogenetic tree of predicted β-lactamases identified. Predicted protein sequences were obtained using ORFfinder (NCBI) (≥50 amino acids in length) using the genes received from functional metagenomics sequencing. β-lactamases (n = 64) were assigned to an Ambler class according the best hit from blast against NCBI. Sequences with less than 85% identity over 90% of sequence to known β-lactamase were classified as non-described proteins. All sequences were aligned with MUSCLE (software MEGA, 5.2.1) and an approximately-maximum-likelihood phylogenetic tree was built using FastTree (http://www.microbesonline.org/fasttree/, July 2017).
