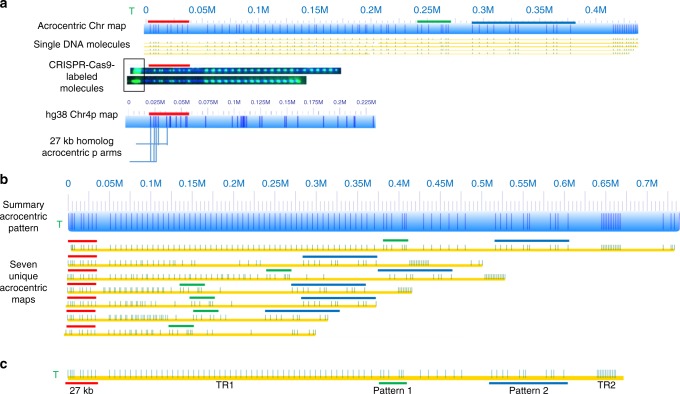
Fig. 7.
Acrocentric chromosome patterns in non-aligned maps. Non-aligned maps were grouped into unique patterns. These groups were analyzed for patterns localizing maps to near telomeric regions. a An acrocentric map (blue) supported by molecules (yellow) with Nt.BspQI labels (green dots) aligned to an in silico labeled map (dark blue vertical lines). Molecules comprising the maps were localized to acrocentric regions by a CRISPR-Cas9 labeling method described previously. The green T indicates telomere labeling. The 27-kb homolog, shown by the blue teeth underneath the chromosome 4p map, was identified by in silico nicking of the chromosome 4p sequence indicated by red bars throughout the figure. b Seven unique acrocentric maps (yellow bars with green BspQI labels) were identified and aligned to a summarized version (blue bar). The red-, green-, and blue-colored bars indicate the elements defining an acrocentric map. c A general model for an expected acrocentric map. From the telomeric end (green T), the 27-kb homolog must be present (red bar), followed by tandem repeat region 1 (TR1), a unique labeling pattern (green bar, Pattern 1), a second unique labeling pattern (blue bar, Pattern 2), and a final tandem repeat region (TR2). Patterns 1 and 2 exhibit little variation, while unit counts of the tandem repeat regions varied considerably