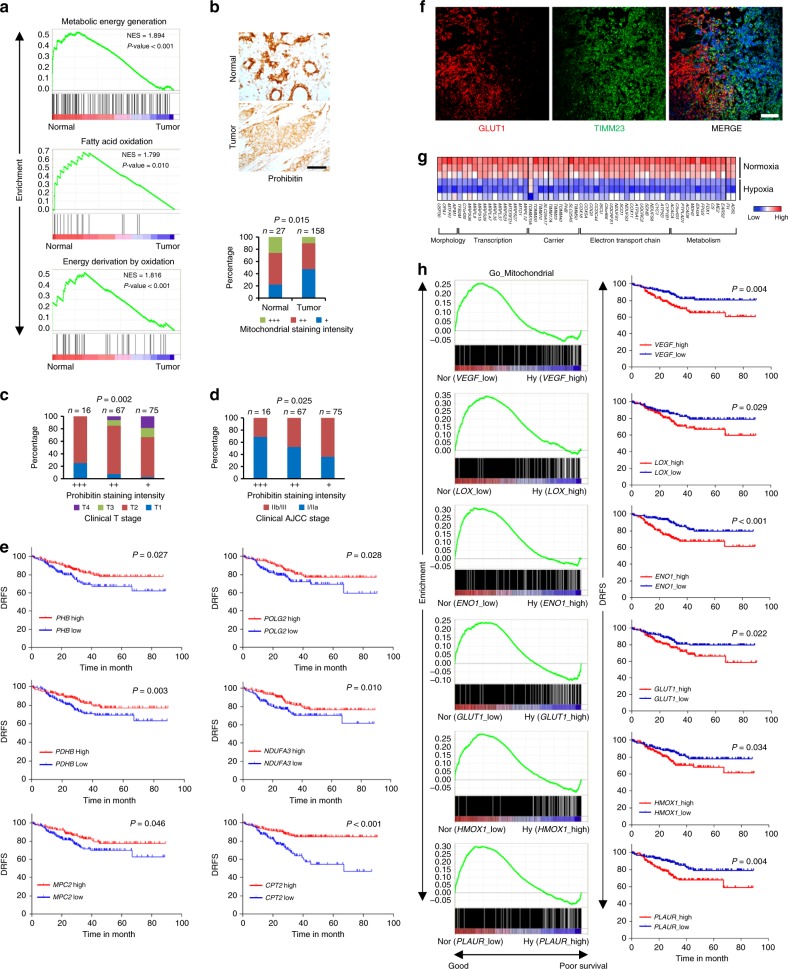
Fig. 1.
Nuclear-encoded mitochondrial gene expression negatively correlates with clinical outcome in breast cancer patients. a GO (Gene Ontology) enrichment analysis in normal and breast cancer tissues from dataset GSE15852, with a false discovery rate (FDR) of <25%. b Top: immunohistochemical staining of Prohibitin in a representative normal breast tissue sample and breast cancer tissue sample from the tissue microarray. Scale bars, 50 μm. Brown color indicates positive immune reaction. Bottom: graph showing the intensity of Prohibitin staining in 27 normal and 158 tumor tissue samples. Statistical significance was determined by χ2 test. c, d Statistical analysis of correlations between Prohibitin staining intensity and clinical T stage (c) or clinical AJCC stage (d). Statistical significance was determined by χ2 test. e Analysis of the correlations between disease relapse-free survival rate (DRFS) and the expressions of indicated genes in breast cancer patients from dataset GSE25055. f Mouse spontaneous breast tumor tissues were stained with anti-GLUT1 (red) and anti-TIMM23 (green) antibodies together with DAPI (blue). Scale bars, 50 µm. g Heat map of the expression of mitochondrial genes in MDA-MB-231 cells cultured under normoxia and hypoxia (dataset GSE18494). h Analysis of the correlations between DRFS and GO enrichment in mitochondrial genes in breast cancer patients from dataset GSE25055