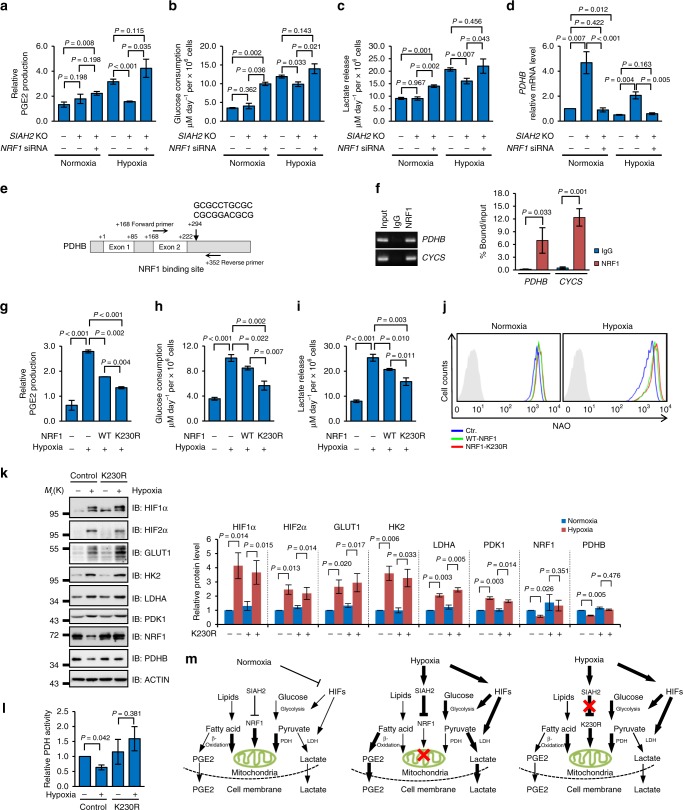
Fig. 6.
The SIAH2-NRF1 axis facilitates hypoxia-induced metabolic reprogramming. a–d Wild-type, SIAH2−/− and SIAH2−/−/NRF1 siRNA MDA-MB-231 cells were cultured under normoxia or hypoxia for 36 h, and concentrations of prostaglandin E2 (PGE2) within cells (a), glucose (b), lactate (c) in the culture medium and mRNA levels of PDHB (d) were analyzed. qRT-PCR results were normalized to the housekeeping gene B2M. e Diagram showing the NRF1 binding site in PDHB gene and oligonucleotides used in the ChIP assay. f ChIP assay was performed with IgG and antibody against NRF1 and indicated genes were analyzed by qRT-PCR. qRT-PCR results were normalized to the input. g–i MDA-MB-231 cells, either mock-treated or stably expressing wild-type NRF1 or the NRF1-K230R mutant, were cultured under normoxia or hypoxia for 36 h, and concentrations of prostaglandin E2 (PGE2) (g), glucose (h) and lactate (i) were analyzed. j Indicated cells were cultured under normoxia or hypoxia for 36 h, cells were stained with 100 nM Nonyl Acrdine Orange (NAO) and analyzed. k Wild-type or K230R stably expressed MDA-MB-231 cells were cultured under normoxia or hypoxia for 36 h. Cells were harvested and analyzed by western blotting with indicated antibodies. Right: densitometric quantification of the indicated proteins. l Cells from (k) were collected and PDH activity was measured. m A proposed model of the SIAH2-NRF1 axis in regulating hypoxia-induced metabolic reprogramming. For all panels, error bars indicate s.d., n = 3 biological replicates, average of n = 3 technical replicates for each biological replicate was used in (a–d), (f–i) and (l). The two-tailed unpaired student t-test was used in (a–c) and (f–i). Two-tailed paired ratio t-test was used in (d) and (k–l)