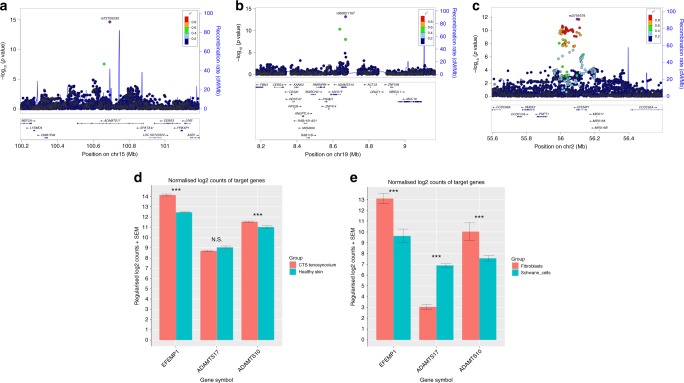
Fig. 2.
Regional association plots for three GWAS loci and the expression of target genes from RNA-Seq. a Chromosome 15q26.3 (the site of missense variant in ADAMTS17). b Chromosome 19p13.2 (the site of missense variant in ADAMTS10). c Chromosome 2p16.1 (the site of intronic variant in EFEMP1). SNP position is shown on the x-axis, and the strength of association on the y-axis. The linkage disequilibrium (LD) relationship between the lead SNP and the surrounding SNPs is indicated by the colour. In the lower panel of each figure, genes within 500 kb of the index SNP are shown. The position on each chromosome is shown in relation to Human Genome build hg19. d Comparison of gene expression between tenosynovium of 41 CTS cases and the index finger skin of six healthy individuals. e Comparison of gene expression between human cultured fibroblasts and Schwann cells from Weiss et al.38. Error bars represent the standard error of the mean of the regularised log2 counts. p Value was determined using Wald test and was FDR adjusted. ***p Value < 0.01; NS = not significant. Source data are provided as a Source Data file. CTS, carpal tunnel syndrome; FDR, false discovery rate; GWAS, genome-wide association study; RNA-Seq, RNA sequencing; SNP, single-nucleotide polymorphism