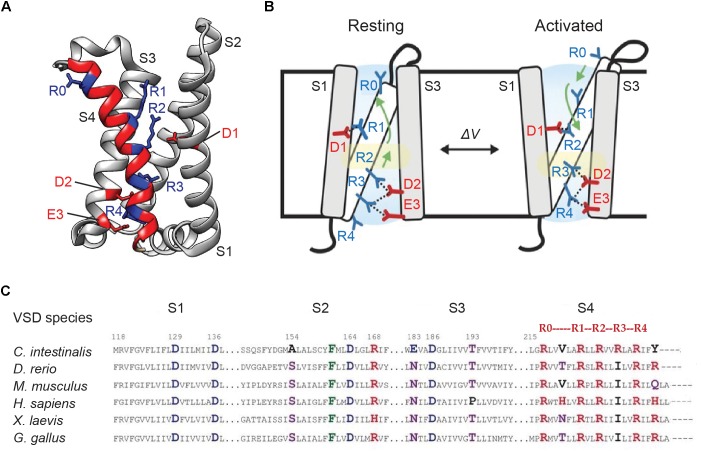
FIGURE 5.
Genetically encoded voltage indicators kinetics and voltage dependence of optical responses largely depend on voltage-sensitive conformational changes in its VSD. (A) Crystal structure of the VSD of Ciona intestinalis voltage sensitive phosphatase (PDB ID 4G7V). The four transmembrane helices S1–S4 are shown. S4 is highlighted in red. The position of the gating arginines R1–R4 (every third amino acid starting at 223) as well as the outermost S4 arginine R0 (position 217 in CiVSD) are shown in blue. The countercharges D1 (on S2 helix), D2 and E3 (on S3 helix) are indicated in red. (B) A mechanistic model of voltage sensing in CiVSD. The activated UP state is accompanied by a 5 Å S4 down movement and a ∼60° counterclockwise rotation of the entire helix. The arginines are then stabilized by successive negative countercharges, from R1–D1, R2–D2, and R3–D3 pairings in the resting or DOWN state to R2–D1, R3–D2, and R4–E3 in the UP configuration. (C) Sequence alignment of the VSD transmembrane segments of voltage sensitive phosphatase orthologs. Positive, negative and polar residues are indicated in red, blue, and purple, respectively. The numbering follows that of the Ciona homolog (Li et al., 2014; Piao et al., 2015).