Fig. 4.
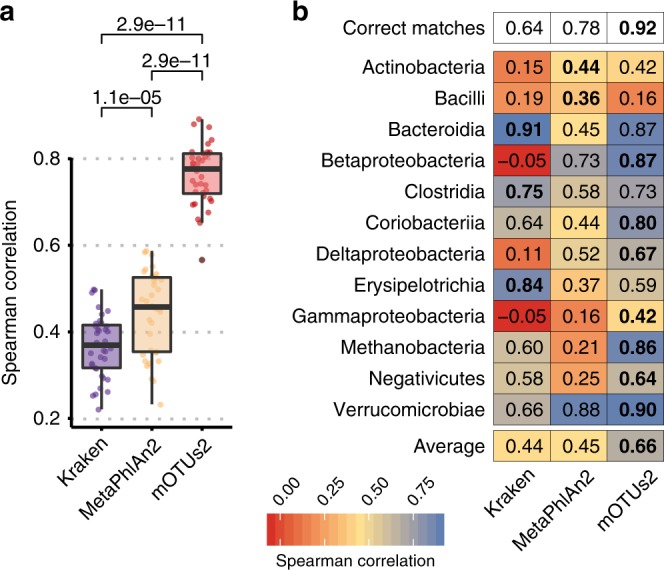
Metatranscriptomic abundance profiling with mOTUs2. a Spearman correlation between matched metagenomic and metatranscriptomic profiles obtained from 36 faecal samples with Kraken, MetaPhlAn2 and mOTUs2. mOTUs2 profiles (red) show significantly higher correlation than the other two methods (paired two-sided Wilcoxon test, boxplots show the median correlation as horizontal lines and interquartile ranges as boxes, whiskers extend at most 1.5 times the interquartile range). b The top-row represents the proportion of cases in which the distance (log-Euclidean) between metagenomic and metatranscriptomic profiles was smallest for the same sample. Below is a taxonomic breakdown (12 most abundant classes) of correlations between metagenomic and metatranscriptomic profiles. For each class, the highest correlation value across the tested methods are shown in bold. Source data are provided as a Source Data file.
