Abstract
Sphingolipid and cholesterol metabolism are closely associated at the structural, biochemical, and functional levels. Although HDL-associated sphingosine-1-phosphate (S1P) contributes to several HDL functions, and S1P signaling regulates glucose and lipid metabolism, no study has addressed the involvement of S1P in cholesterol efflux. Here, we show that sphingosine kinase (Sphk) activity was induced by the LXR agonist 22(R)-hydroxycholesterol and required for the stimulation of ABCA1-mediated cholesterol efflux to apolipoprotein A-I. In support, pharmacological Sphk inhibition and Sphk2 but not Sphk1 deficiency abrogated efflux. The involved mechanism included stimulation of both transcriptional and functional ABCA1 regulatory pathways and depended for the latter on the S1P receptor 3 (S1P3). Accordingly, S1P3-deficient macrophages were resistant to 22(R)-hydroxycholesterol-stimulated cholesterol efflux. The inability of excess exogenous S1P to further increase efflux was consistent with tonic S1P3 signaling by a pool of constitutively generated Sphk-derived S1P dynamically regulating cholesterol efflux. In summary, we have established S1P as a previously unrecognized intermediate in LXR-stimulated ABCA1-mediated cholesterol efflux and identified S1P/S1P3 signaling as a positive-feedback regulator of cholesterol efflux. This constitutes a novel regulatory mechanism of cholesterol efflux by sphingolipids.
Keywords: ATP binding cassette transporter A1, apolipoprotein A-I, atherosclerosis, high density lipoprotein, reverse cholesterol transport, sphingosine-1-phosphate, S1P receptor 3, sphingosine kinases
Atherosclerosis is a chronic inflammatory disease characterized by deposition of excess cholesterol in the arterial circulation and its pathophysiological sequelae and underlies CVDs as the leading cause of death in industrialized nations (1). Reverse cholesterol transport (RCT) is a mechanism by which excess cholesterol from peripheral tissues is taken up by macrophages and transferred to HDLs to be transported to the liver for excretion and is therefore considered a preventive measure in the pathogenesis of atherosclerosis (2). The efflux of cholesterol to HDL particles of different maturation stages is mediated by various transport proteins. The ABCA1 and ABCG1 are responsible for cholesterol efflux to lipid-poor HDL apolipoproteins and ApoA-I and to mature HDLs, respectively (3–5). ABCA1 and ABCG1 function and expression are under the control of complex transcriptional and posttranslational mechanisms. Abca1 transcription is regulated by LXRα, and LXRα ligands such as the oxysterol 22(R)-hydroxycholesterol [22(R)-OHC] potently induce Abca1 and Abcg1 gene expression and cholesterol efflux (6–8). In addition, cAMP acting through cAMP-dependent protein kinase A (PKA) is also a major stimulator of ABCA1 gene and protein expression, and PKA also stimulates ABCA1 function by direct phosphorylation (9, 10). Many inflammatory cytokines, proteins, and lipids influence ABCA1-regulated cholesterol efflux through numerous transcriptional and other regulatory mechanisms (6).
HDLs are highly complex particles of different subclasses, maturation stages, and compositions. Although their main physiological function in vivo is RCT, HDLs have numerous RCT-independent vasoactive, antioxidative, antiinflammatory, and antithrombotic properties (11, 12). Some of these properties are mediated in part by HDL-contained sphingosine-1-phosphate (S1P) (12, 13), a bioactive sphingolipid with fundamental cardiovascular, inflammatory, and immune functions (12, 14). In particular, HDL-associated S1P has been implicated in the regulation of arterial vasomotor tone, endothelial barrier function, angiogenesis, cardioprotection, inflammation, lipid oxidation, and cell survival, with different S1P receptors playing unique, coordinated, or even opposing roles (12). Recently, engineered S1P chaperones based on apolipoprotein M, the main S1P-binding protein in HDL, have been shown to have beneficial effects in experimental models of hypertension, myocardial infarction (MI), neuroinflammation, and stroke (15). However, the involvement of HDL-S1P or S1P on RCT or cholesterol efflux has not been studied. This is somewhat surprising, as there is ample evidence of an involvement of S1P signaling in the regulation of lipid and glucose metabolism at the cellular, organ, and systemic levels, whereas disturbances in S1P homeostasis and signaling have been described in various metabolic disorders (16–18). Furthermore, it is well established that sphingolipid and cholesterol homeostasis and metabolism are closely associated at the structural, biochemical, and functional levels (19–22).
Thus, the aim of the current study was to analyze whether there is a connection between S1P signaling and cholesterol efflux to ApoA-I in murine macrophages. We uncovered an intricate relationship between endogenous S1P synthesis by sphingosine kinases (Sphks), S1P receptor signaling, and ABCA1-mediated cholesterol efflux that constitutes a novel regulatory pathway of HDL function with potential impact on cholesterol homeostasis and RCT.
MATERIALS AND METHODS
Mice
C57Bl6J and floxed ABCA1/G1 mice were obtained from the Jackson Laboratory. S1P1flox/flox, S1P2-deficient, and S1P3-deficient mice were kindly provided by Jerold Chun, Scripps Research Institute (to B.L.). Mice with hematopoietic deletion of S1P1 were generated by cross-breeding S1P1flox/flox mice with mice heterozygous for the Cre recombinase under the control of the vav promoter. Sphk1- and Sphk2-deficient mice were kindly provided by Richard Proia, the National Institutes of Health (to M.H.G.).
Isolation and culturing of peritoneal macrophages
Peritoneal macrophages were isolated 96 h after i.p. injection of sterile aged 3% thioglycolate (Becton Dickinson). To harvest macrophages, 4 ml of prewarmed PBS (AppliChem) was injected into the peritoneum and aspirated. After centrifugation for 5 min at 400 g and 4°C, red blood cells were lysed using ACK buffer (0.15 M NH4Cl, 1 mM KHCO3, and 0.1 mM Na2EDTA; pH 7.2–7.4) for 5 min at room temperature, washed once with ice-cold PBS, counted, and plated. Cultured macrophages were used for different experiements as described below.
Gene expression analysis
For gene expression studies, cells were cultured in RPMI 1640 medium (Invitrogen) containing 1% FBS (Gibco) and 1% antibiotic-antimycotic (PSA: 10,000 U/ml penicillin, 10,000 µg/ml streptomycin, and 25 µg/ml amphotericin B; Gibco). After 2 h, adherent cells were washed with PBS, and the medium was changed to RPMI without FBS. Peritoneal macrophages were treated with 6.25 µM 22(R)-OHC (Sigma-Aldrich) in combination with 1 µM 9-cis-retinoic acid (9cRA; Sigma-Aldrich), for 18 h at 37°C. The Sphk inhibitor 4-{[4-(4-chlorophenyl)-2-thiazolyl]amino}-phenol (SKi) (Cayman Chemical) was used at 5 µM as indicated. Total RNA was isolated using the InnuPrep RNA Mini kit from Analytic Jena according to the manufacturer’s protocol. cDNA was generated from 100 ng of RNA with the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, Germany). Quantitative real-time PCR was performed in a C1000TM Thermal Cycler CFX96 using iQ SYBR Green Supermix (Bio-Rad) and a standard RT-PCR program was used: initial denaturation for 3 min at 95°C, followed by 40 cycles of 10 s denaturation at 95°C, 10 s annealing at 55°C, and 30 s elongation at 72°C, followed by 10 min at 95°C and the melting curve of incremental increases of 0.5°C every 5 s from 65°C to 95°C. Relative quantification of gene expression was calculated by the 2-ΔΔCT method using Gapdh as an endogenous reference for normalization. Abca1, Gapdh, and Sphk2 primers were ordered from BioTez, and Sphk1 primers were ordered from Qiagen (Table 1).
TABLE 1.
Primer sequences used for quantitative real-time PCR
| Mouse Genes | Primer Sequence (5′–3′) |
| Abca1 | For: AAG GAA GTT GGC AAG GTT GG |
| Rev: CCA TGC CTG TGG TTG GTT CA | |
| Gapdh | For: AGG TCG GTG TGA ACG GAT TTG |
| Rev: TGT AGA CCA TGT AGT TGA GGT CA | |
| Sphk1 | Qiagen catalog no. QT01046395 |
| Sphk2 | For: CAC GGC GAG TTT GGT TCC TA |
| Rev: CTT CTG GCT TTG GGC GTA GT |
For., forward; Rev., reverse.
Cholesterol efflux assay
Peritoneal macrophages from all indicated mouse strains were plated in RPMI 1640 medium supplemented with 1% FBS and 100 U/ml penicillin/100 µg/ml streptomycin (Invitrogen) and allowed to adhere for 2 h at 37°C and 5% CO2. After washing twice with prewarmed PBS, macrophages were loaded with 1 μCi/ml 3H-cholesterol (PerkinElmer) in RPMI containing 1% FBS and 2 µg/ml acyl-CoA cholesterol acyltransferase inhibitor (ACATi; Sandoz 58-035, Santa Cruz) for 24 h. The following day, the cells were washed once with PBS and supplemented with 200 µl of RPMI 1640, containing ACATi without FBS. For stimulation, 5 µM SKi, 6.25 µM 22(R)-OHC, 1 µM 9cRA, and 1 µM S1P (Enzo) either alone or in combination were added, and the macrophages were then incubated for 18 h at 37°C. After 18 h of treatment, the cells were washed twice with prewarmed PBS. Efflux medium (αMEM with 25 mM HEPES; Invitrogen) supplemented with 2 µg/ml ACATi and 10 µg/ml human plasma-derived ApoA-I (Acris) was added for 5 h. Stimulation with 1 µM S1P or inhibition with 5 µM SKi was performed by adding S1P/SKi 30 min before adding the ApoA-I. After 5 h, the medium was transferred to 1.5 ml tubes. Following centrifugation (5 min, 300 g at room temperature) to pellet cellular debris, radioactivity in the supernatant was determined by liquid scintillation counting (PerkinElmer). Macrophages were washed once with PBS, and intracellular lipid extraction was performed by adding 200 µl hexane:isopropanol (3:2, vol:vol) (Applichem/Roth) for 30 min at room temperature. The hexane/isopropanol fraction was transferred into a 1.5 ml tube. Hexane/isopropanol was added again to the cells, incubated for another 15 min at room temperature, and transferred to the same tube. The tubes were allowed to dry overnight under the hood in order to get rid of the hexane/isopropanol. The following day, the extracted lipids were dissolved in isopropanol:NP-40 (9:1, vol:vol) (Roth/Fluka Biochemika) and subjected to scintillation analysis. Efflux per well was expressed as the percentage of counts released into the medium relative to the total amount of radioactivity initially present (counts recovered in the medium added to the counts recovered from the cells). Values obtained from control cells without added ApoA-I were subtracted from all respective experimental values to correct for baseline efflux.
C17-S1P assay
Peritoneal macrophages from C57Bl/6 mice were plated at a density of 2 × 106 cells/well in a 6-well plate using RPMI 1640 medium supplemented with 1% FBS and 100 U/ml penicillin/100 µg/ml streptomycin (Invitrogen) and allowed to adhere for 2 h at 37°C and 5% CO2. After washing with prewarmed sterile PBS, cells were treated with 6.25 µM 22(R)-OHC and 1 µM 9cRA in presence or absence of 5 µM SKi in RPMI 1640 without FBS for 18 h. For the last hour, cells were spiked with 10 µM C17-sphingosine (Avanti), and at the end, the supernatants were harvested for measurement of C17-S1P by LC-MS/MS.
Sphk activity assay
Enzyme assays were performed as previously described (23). In brief, 10 μg of total protein lysate (prepared as for Western blotting) was incubated with 100 µM sphingosine and 1 μCi [γ-32P]-ATP (3,000 Ci/mmol, Perkin Elmer) in 1 mM ATP in assay buffer [100 mM Tris-HCl (pH 7.4), 0.025% TX-100, 200 mM MgCl2, 1 mM Na3VO4 (Sigma-Aldrich), 10 mM NaF (Sigma-Aldrich), and 0.5 mM 4-deoxypyridoxine (Sigma-Aldrich)] in a total volume of 100 μl for 30 min at 37°C. The enzyme reaction was terminated by addition of 270 μl of chloroform:methanol:concentrated HCl (100:200:1). S1P was extracted by the addition of 50 µ 2 M KCL and 70 µl chloroform to create a phase separation (24). After centrifugation for 5 min at 13,000 g, the upper phase was removed by aspiration. Labeled S1P in the lower phase was quantified by TLC. Briefly, 40 µl lipid samples were spotted onto TLC Silicagel 60 F254 (Merck Millipore) and resolved using l-butanol/ethanol/acetic acid/water at (8/2/1/2) as the development system. Resolved lipids were visualized using storage phosphor screens processed on a Typhoon phosphoimager with ImageQuant software. 32P-S1P was located based on comigration with a prepared S1P standard. To quantify 32P-S1P, known quantities of radiolabeled ATP were spotted onto each plate to generate standard curves, from which the amounts of ATP incorporated into 32P-S1P were determined.
Western blotting
For Western blotting, peritoneal macrophages were isolated as stated before. After 2 h, adherent cells were treated with 6.25 µM 22(R)-OHC and 1 µM 9cRA in the absence of presence of 5 µM SKi in RPMI for 18 h. Cells were then homogenized in RIPA buffer, protein concentration was determined with the BCA assay (Pierce), and 40 µg of protein was run on SDS-PAGE, transferred to Immobilon-P PVDF membrane (Merck Millipore), and blotted with Abs to ABCA1 (Novus Biologicals, catalog no. NB400-105) and vinculin (Chemicon, catalog no. MAB1624). The electrochemiluminescence signal was quantified by a Bio-Rad ChemiDoc MP Imaging System using Image Lab 3.0.1 (Beta 2).
Statistical analysis
Each experiment was performed at least three times. Statistical significance was evaluated by either one-way ANOVA, two-way ANOVA, or a two-tailed paired or unpaired Student’s t-test, whichever was appropriate, and as indicated in each figure legend. P values of less than 0.05 were considered statistically significant. The graphs were generated using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA). The data are presented as mean ± SEM.
RESULTS
Pharmacological and genetic blockade of Sphk 2 inhibits 22(R)-OHC-stimulated cholesterol efflux in macrophages
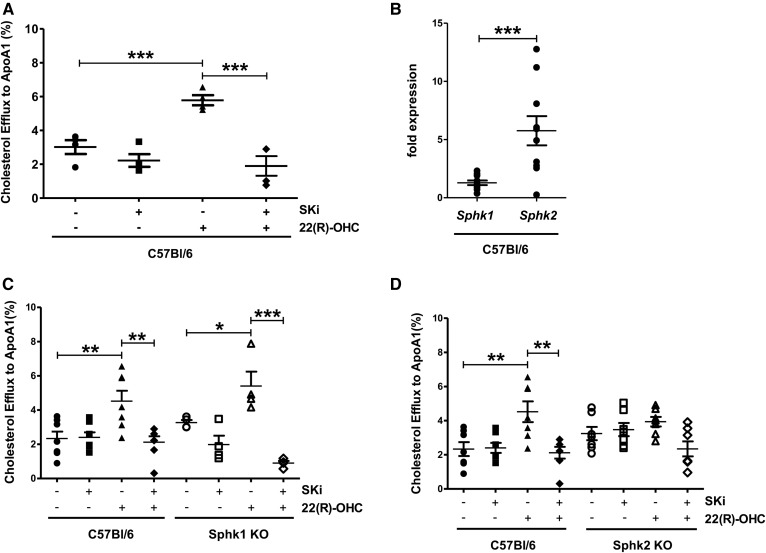
To examine the role of Sphks on cholesterol efflux to ApoA-I in C57Bl6J peritoneal mouse macrophages, we stimulated ABCA1-mediated cholesterol efflux by the LXR agonist 22(R)-OHC in the presence and absence of the dual Sphk1 and Sphk2 inhibitor 4-{[4-(4-chlorophenyl)-2-thiazolyl]amino}phenol (SKi) (25–27). SKi has been shown to inhibit efficiently both enzymes (26). We observed that SKi did not affect basal but completely abrogated 22(R)-OHC-stimulated cholesterol efflux (Fig. 1A). In macrophages, gene expression of Sphk2 was 5-fold higher than Sphk1 (Fig. 1B), consistent with previous reports (28). To differentiate between the role of Sphk1 and Sphk2 in cholesterol efflux and corroborate the pharmacological findings by genetic means, we analyzed cholesterol efflux in Sphk1- and Sphk2-deficient macrophages, respectively. Although 22(R)-OHC-stimulated efflux did not differ between Sphk1-deficient and WT macrophages (Fig. 1C), it was entirely abrogated in Sphk2-deficient macrophages (Fig. 1D). In line with a causal role of Sphk2, SKi inhibited 22(R)-OHC-stimulated cholesterol efflux in Sphk1-deficient but not Sphk2-deficient macrophages (Fig. 1C, D).
Fig. 1.
Pharmacological inhibition and genetic deletion of Sphk2 inhibits 22(R)-OHC-stimulated cholesterol efflux to ApoA-I. A: 3H-cholesterol efflux to ApoA-I was measured in C57Bl6 macrophages incubated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 4 per group). Statistical analysis was performed using one-way ANOVA followed by a Student-Newman-Keuls test. *** P < 0.001. B: Sphk1 and Sphk2 gene expression was analyzed by quantitative real-time PCR in C57BL6 macrophages. Individual points represent individual mice. Data are presented as mean ± SEM (n = 10 per group). Statistical analysis was performed using a two-tailed paired Student’s t-test. *** P < 0.001. C: 3H-cholesterol efflux to ApoA-I was measured in Sphk1 KO and WT macrophages treated as in A. Individual points represent individual mice. Data are presented as mean ± SEM (n = 4–7 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. * P < 0.05; ** P < 0.01; *** P < 0.001. D: 3H-cholesterol efflux to ApoA-I was measured in Sphk2 KO and WT macrophages treated as in A. Individual points represent individual mice. Data are presented as mean ± SEM (n = 7 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. ** P < 0.01.
Sphk inhibition abrogates 22(R)-OHC-stimulated cholesterol efflux by blocking Abca1 transcription
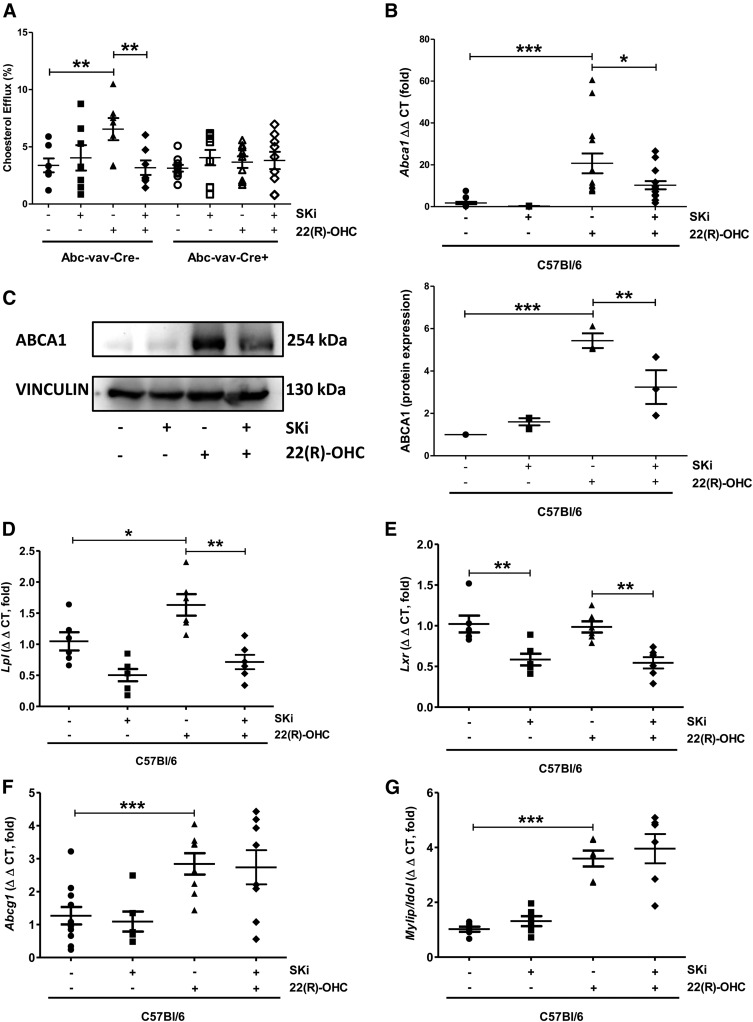
ABCA1 is transcriptionally regulated by LXR and is responsible for the cholesterol efflux to ApoA-I. We thus tested whether SKi affected cholesterol efflux in an ABCA1-dependent manner. Indeed, no induction of cholesterol efflux by 22(R)-OHC was observed in macrophages lacking ABCA1, and SKi displayed no inhibitory effect beyond that caused by ABCA1 deficiency (Fig. 2A). These data strongly argue for a requirement of Sphk activity for 22(R)-OHC-induced ABCA1-mediated cholesterol efflux. As 22(R)-OHC stimulates cholesterol efflux to ApoA-I by inducing LXR-dependent Abca1 gene transcription, we analyzed whether the inhibitory effect of SKi was due to effects on Abca1 transcription. Indeed, SKi greatly suppressed ABCA1 gene and protein expression (Fig. 2B, C). This was also the case for other LXR-dependent genes, such as Lpl (lipoprotein lipase) and Lxr itself (Fig. 2D, E) but not Abcg1 and Mylip/Idol (Fig. 2F, G).
Fig. 2.
ABCA1 is required for the effect of Sphk inhibition on 22(R)-OHC-stimulated cholesterol efflux. A: 3H-cholesterol efflux to ApoA-I was measured in ABCA1 KO and WT macrophages incubated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 7–10 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. ** P < 0.01. B: Abca1 gene expression was analyzed by quantitative real-time PCR in C57BL6 macrophages treated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 10–12 per group). Statistical analysis was performed using one-way ANOVA followed by a Student-Newman-Keuls test. * P < 0.05; *** P < 0.001. C: ABCA1 protein expression was analyzed by Western blotting in C57BL6 macrophages treated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h. Quantification was performed in relation to vinculin. Statistical analysis was performed using one-way ANOVA followed by a Student-Newman-Keuls test. ** P < 0.01; *** P < 0.001. D–G: Gene expression of Lpl, LXRα, Abcg1, and Mylip/Idol was analyzed by real-time PCR in C57BL6 macrophages treated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 6–11 per group). Statistical analysis was performed using a two-tailed paired Student’s t-test. * P < 0.05; ** P < 0.01; *** P < 0.001.
22(R)-OHC stimulates Sphk activity
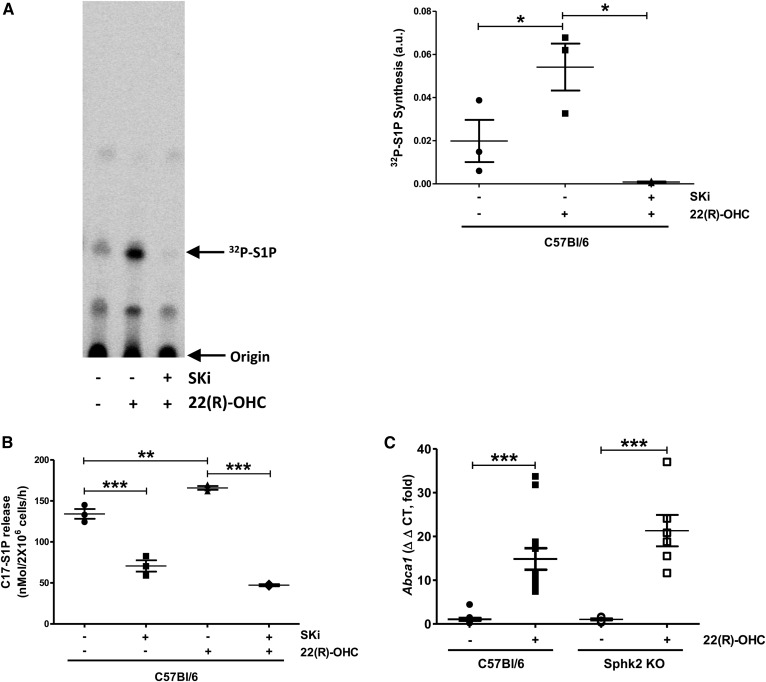
To understand the relationship between Sphk activity and LXR-regulated ABCA1-mediated cholesterol efflux, we tested whether 22(R)-OHC induced Sphk activity. To do this, we used two different approaches. In the first, we performed Sphk kinase assays using 32P-labeled ATP as described (23). We observed that 22(R)-OHC greatly stimulated Sphk activity and that SKi completely abolished this stimulation (Fig. 3A). In a second approach, we measured generation and release of C17-S1P from intact cells by LC/MS/MS from C17-sphingosine added to the cells for the final hour of the 18 h 22(R)-OHC stimulation period. As with the in vitro assay, 22(R)-OHC stimulated Sphk activity as mirrored by the release of C17-S1P release, and this was reliably inhibited by SKi (Fig. 3B). This suggested that LXR agonism stimulated Sphk activity and that the latter was required for LXR-dependent Abca1 transcription. Surprisingly, despite the clear inhibition of 22(R)-OHC-mediated cholesterol efflux by SKi in WT and abrogation in Sphk2 KO macrophages, 22(R)-OHC was still able to induce Abca1 gene expression in Sphk2-KO macrophages (Fig. 3C). This suggests posttranscriptional in addition to transcriptional mechanisms by which Sphk2 influenced ABCA1 function.
Fig. 3.
The 22(R)-OHC stimulates Sphk activity and S1P release in C57Bl/6 macrophages. A: Sphk activity was measured in lysates from peritoneal macrophages treated with 6.25 μM 22(R)-OHC for 18 h. Individual points represent experiments with individual mice. Data are presented as mean ± SEM (n = 3 per group). Statistical analysis was performed using a two-tailed paired Student’s t-test. * P < 0.05. B: C17-S1P released from macrophages treated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h and incubated with C17-sphingosine in the last hour was measured by mass spectrometry. Individual points represent individual mice. Data are presented as mean ± SEM (n = 3 per group). Statistical analysis was performed using one-way ANOVA followed by a Student-Newman-Keuls test. ** P < 0.01; *** P < 0.001. C: Abca1 gene expression was analyzed by quantitative real-time PCR in Sphk2 KO and WT macrophages treated with 6.25 μM 22(R)-OHC for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 6–13 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. *** P < 0.001.
Role of S1P receptors in 22(R)-OHC-stimulated cholesterol efflux
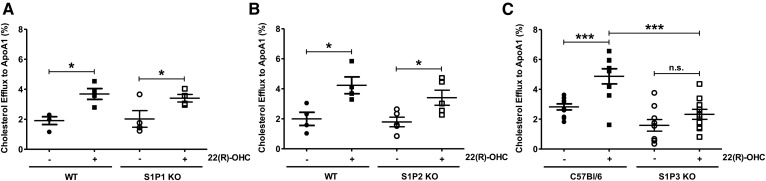
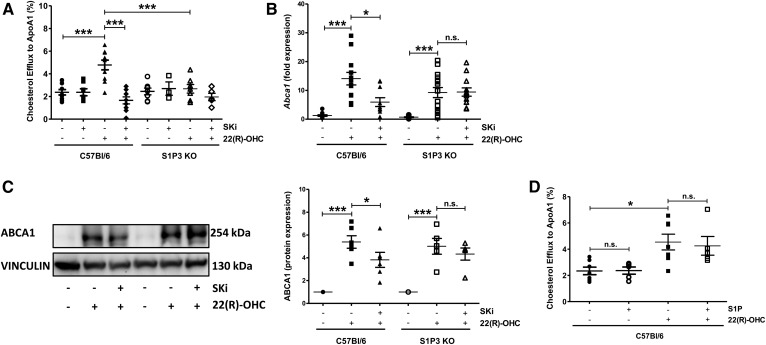
To test whether Sphk activity exerted its effect on ABCA1 by activating S1P receptors, we analyzed basal and 22(R)-OHC-stimulated cholesterol efflux in S1P receptor 1 (S1P1)-deficient, S1P receptor 2 (S1P2)-deficient, and S1P receptor 3 (S1P3)-deficient macrophages. In the absence of S1P1 or S1P2, basal and 22(R)-OHC-stimulated cholesterol efflux was comparable to that of WT macrophages (Fig. 4A, B). In contrast, 22(R)-OHC was unable to stimulate cholesterol efflux in S1P3-deficient macrophages (Fig. 4C), where SKi had no additional inhibitory effect (Fig. 5A). This observation suggested that the stimulatory Sphk effect on 22(R)-OHC-mediated cholesterol efflux was exercised by the S1P3 receptor. As 22(R)-OHC was unable to stimulate Abca1 transcription in the presence of SKi, we assumed that this would be also the case in S1P3-deficient macrophages. However, despite the clear lack of 22(R)-OHC-stimulated efflux in S1P3-deficient macrophages, 22(R)-OHC stimulated Abca1 gene expression as efficiently as in WT macrophages (Fig. 5B). Another surprise was that SKi did not suppress 22(R)-OHC-stimulated ABCA1 gene and protein expression in S1P3-deficient macrophages (Fig. 5C). These observations suggested that 22(R)-OHC-stimulated cholesterol efflux required Sphk activation and intact S1P3 signaling but that the mere lack of S1P3 already prevented cholesterol efflux through posttranscriptional ABCA1 inhibition. Finally, we tested for additional effects of exogenous S1P on 22(R)-OHC-stimulated cholesterol efflux but observed none (Fig. 5D).
Fig. 4.
The 22(R)-OHC is unable to stimulate cholesterol efflux in S1P3-deficient macrophages. A: 3H-cholesterol efflux to ApoA-I was measured in S1P1 KO and WT macrophages, incubated with 6.25 μM 22(R)-OHC for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 4 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. * P < 0.05. B: 3H-cholesterol efflux to ApoA-I was measured in S1P2 KO and WT macrophages treated as in A. Individual points represent individual mice. Data are presented as mean ± SEM (n = 4 or 5 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. * P < 0.05. C: 3H-cholesterol efflux to ApoA-I was measured in S1P3 KO and WT macrophages, incubated as in A. Individual points represent individual mice. Data are presented as mean ± SEM (n = 9 or 10 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. *** P < 0.001. n.s., not significant.
Fig. 5.
Lack of 22(R)-OHC-stimulated-cholesterol efflux in S1P3-deficient macrophages. A: 3H-cholesterol efflux to ApoA-I was measured in S1P3 KO and WT macrophages incubated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 3–10 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. *** P < 0.001. B: Abca1 gene expression was analyzed by real-time PCR in S1P3 KO and WT macrophages treated with 6.25 μM 22(R)-OHC in the presence and absence of 5 μM SKi for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 10–15 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. * P < 0.05; *** P < 0.001. C: ABCA1 protein expression was analyzed by Western blotting in S1P3 KO and WT macrophages treated as in B. Quantification was performed in relation to vinculin. Individual points represent individual mice. Data are presented as mean ± SEM (n = 5 or 6 per group). Statistical analysis was performed using two-way ANOVA followed by Bonferroni test. * P < 0.05; *** P < 0.001. D: 3H-cholesterol efflux to ApoA-I was measured in C57Bl6 macrophages incubated with 6.25 μM 22(R)-OHC in the presence and absence of 1 μM S1P for 18 h. Individual points represent individual mice. Data are presented as mean ± SEM (n = 5–7 per group). Statistical analysis was performed using one-way ANOVA followed by a Student-Newman-Keuls test. * P < 0.05. n.s., not significant.
DISCUSSION
This study is the first to identify endogenous S1P generation and signaling as major regulators of ABCA1-mediated cholesterol efflux. The only sphingolipid known so far to affect cholesterol efflux has been ceramide, an S1P precursor, that was shown to increase the plasma membrane presence of ABCA1 and stimulate cholesterol efflux to ApoA-I (29), respectively, but the mechanism has remained unknown. Proof for ABCA1 as the cholesterol transporter affected by S1P in our study stems from the absence of S1P-related effects in ABCA1-deficient macrophages. Our conclusion that Sphk activity played a central role in ABCA1-mediated cholesterol efflux is supported by the data that pharmacological Sphk inhibition and genetic Sphk2 deletion, respectively, suppressed cholesterol efflux after stimulation by the endogenous LXR agonist 22(R)-OHC. Of note, 22(R)-OHC activated Sphk, another novel finding of this study, along with its apparent necessity for the stimulation of cholesterol efflux through transcriptional ABCA1 activation (Fig. 6). These data establish S1P production as a previously unrecognized intermediate in LXR signaling. Some classical LXR-regulated genes such as Lpl were also dependent on Sphk activation, whereas others (Abcg1 and Mylip/Idol) remained unaffected, with the reason for the latter remaining unknown and precluding a generalization of our findings to all LXR-regulated genes.
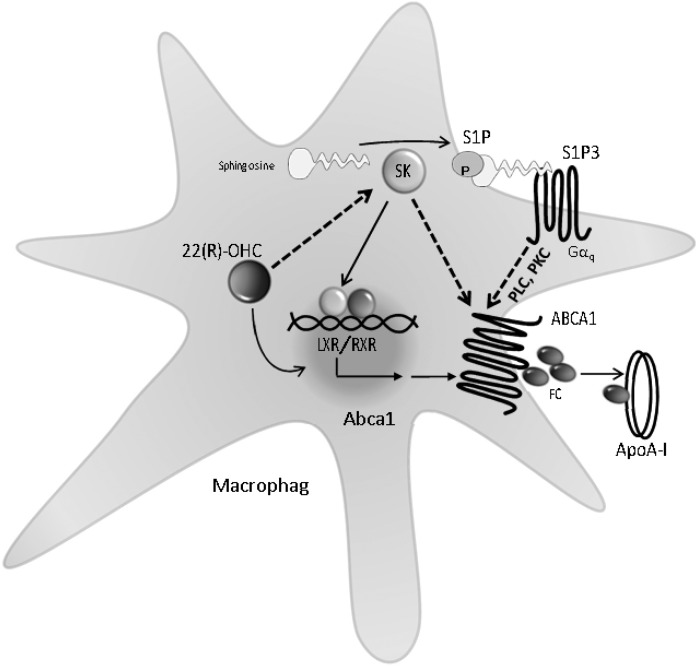
Fig. 6.
Regulation of cholesterol efflux by Sphks, S1P, and S1P3 in macrophages. FC, free cholesterol; Gαq, G protein αq; RXR, retinoid X receptor; SK, Sphks. Dotted lines indicate functional activation, e.g., through yet-uncharacterized posttranslational modifications that result in increased cholesterol efflux.
We favor a hypothesis that there are two simultaneously required mechanisms of how Sphk activation and Sphk-generated S1P affected cholesterol efflux in macrophages (illustrated in Fig. 6): the first mechanism activated Abca1 transcription (not through S1P3, as transcription was unaltered in S1P3-KO), whereas the second stimulated efflux (through S1P3) due to a posttranslational functional activation of ABCA1 (as there was no efflux in S1P3-KO despite intact LXR-stimulated Abca1 transcription). With respect to the first mechanism, 22(R)-OHC-activated Sphk and de novo S1P generation/release and kinase activity was required for efficient efflux. Activation of Sphk by 22(R)-OHC occurred at the posttranscriptional level, as Sphk1 and Sphk2 transcription remained unchanged. How this may have occurred is still unknown. We have identified SPHK2 as the enzyme required for stimulation of cholesterol efflux and the inhibition of Abca1 transcription by SKi. Yet 22(R)-OHC still induced Abca1 gene expression in Sphk2-KO macrophages, suggesting an additional posttranscriptional pathway or a compensatory role for SPHK1. Indeed, Sphks have intracellular targets such as TNF receptor-associated factors, histone deacetylases, and others (30), whereas SKi had an inhibitory, albeit still insignificant, effect in Sphk2-KO macrophages. Furthermore, we have excluded S1P1 and S1P2 as being involved because LXR-stimulated efflux was unaltered in macrophages deficient for the respective receptors. We don’t know whether the less-prominent S1P4 or S1P5 may play a role. As to the second mechanism of a functional ABCA1 activation through S1P3, it may involve stimulation of ABCA1 activity, e.g., through phosphorylation by PKC (31, 32), as S1P3 activates phospholipase C and subsequently PKC through its main coupling to Gαq (30) and PKC has been demonstrated to stimulate cholesterol efflux by phosphorylating and stabilizing ABCA1 (31, 32). We have no data on the latter, but, if true, lack of S1P3 may lead to inefficient cholesterol efflux in the presence of preserved Abca1 gene induction, as observed here.
Interesting, but perhaps not surprising, was the inability of exogenous S1P to stimulate cholesterol efflux regardless of the mean presence of S1P3 being already required for efflux. A probable explanation may lie in a continuously activated, tonic S1P3 signaling by a pool of constitutively generated Sphk-derived S1P as a measure to ensure a steady flow of cholesterol efflux. There is a precedence for such tonic S1P receptor signaling, e.g., in the maintenance of basal Na+/H+ exchanger activity (33) or control of endothelial permeability by erythrocyte-derived S1P (34).
It is well established that ABCA1 mediates not only the efflux of cholesterol but also that of phospholipids to lipid-poor HDL apolipoproteins: intriguingly, ABCA1 has been suggested to act as an S1P exporter in astrocytes in a process involving HDL-like lipoprotein formation (35) and to mediate S1P release to ApoA-I in endothelial cells (36). Thus, a scenario can be envisioned where S1P is exported from macrophages through ABCA1 and regulates cholesterol efflux in a positive S1P3-dependent feedback manner (Fig. 6). The sequence of events need not be simultaneous, as ApoA-I is known to mediate cholesterol and phospholipid transfer in a nonparallel manner (37), so that a truly dynamic regulation of ABCA1-mediated cholesterol efflux by S1P export may be operational and in place. Interestingly, deletion of the S1P-degrading enzyme S1P lyase has been shown to increase plasma lipids and HDL cholesterol (38), suggesting the possibility of such cross-talk. Considering the fact that HDL particles of many different maturation stages carry S1P, HDL-associated may also determine the rate of cholesterol efflux, as is the case for other HDL-S1P-mediated processes (12). Such HDL-S1P-mediated interaction in cholesterol efflux may be pathophysiologically relevant, as HDL-S1P is reduced in atherosclerosis, MI, and diabetes and responsible for such disease-associated impairments of certain HDL functions (11, 12, 39). Interestingly, cholesterol efflux to HDL is compromised in MI and metabolic syndrome (40, 41), suggesting that the low HDL-S1P characteristic for these conditions may be contributing to the defect. As supplementation of S1P-deficient HDL with exogenous S1P can restore some of its compromised functions in disease such as vasodilation and antiinflammation (13, 42), it is plausible to assume that altering plasma S1P or HDL-S1P may be a new approach to regulating cholesterol efflux.
Footnotes
Abbreviations:
- ACATi
- acyl-CoA cholesterol acyltransferase inhibitor
- MI
- myocardial infarction
- RCT
- reverse cholesterol transport
- S1P
- sphingosine-1-phosphate
- S1P1
- S1P receptor 1
- S1P2
- S1P receptor 2
- S1P3
- S1P receptor 3
- Sphk
- sphingosine kinase
- SKi
- sphingosine kinase inhibitor
- 22(R)-OHC
- 22(R)-hydroxycholesterol
- 9cRA
- 9-cis-retinoic acid
This work was supported by Deutsche Forschungsgemeinschaft Grant GRK2098, projects 10 and 11, and Grants SFB 1116 and TPB11; and by an Alexander von Humboldt postdoctoral fellowship (to M.V.). The authors declare that no conflict of interest exists.
REFERENCES
- 1.Lloyd-Jones D., Adams R. J., Brown T. M., Carnethon M., Dai S., De Simone G., Ferguson T. B., Ford E., Furie K., Gillespie C., et al. . 2010. Executive summary: heart disease and stroke statistics—2010 update: a report from the American Heart Association. Circulation. 121: 948–954. [DOI] [PubMed] [Google Scholar]
- 2.Rosenson R. S., Brewer H. B. Jr., Davidson W. S., Fayad Z. A., Fuster V., Goldstein J., Hellerstein M., Jiang X. C., Phillips M. C., Rader D. J., et al. . 2012. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 125: 1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips M. C. 2018. Is ABCA1 a lipid transfer protein? J. Lipid Res. 59: 749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis G. A. 2010. The complexity of HDL. Biochim. Biophys. Acta. 1801: 1286–1293. [DOI] [PubMed] [Google Scholar]
- 5.Cavelier C., Lorenzi I., Rohrer L., and von Eckardstein A.. 2006. Lipid efflux by the ATP-binding cassette transporters ABCA1 and ABCG1. Biochim. Biophys. Acta. 1761: 655–666. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz G., and Langmann T.. 2005. Transcriptional regulatory networks in lipid metabolism control ABCA1 expression. Biochim. Biophys. Acta. 1735: 1–19. [DOI] [PubMed] [Google Scholar]
- 7.Ma Z., Deng C., Hu W., Zhou J., Fan C., Di S., Liu D., Yang Y., and Wang D.. 2017. Liver X receptors and their agonists: targeting for cholesterol homeostasis and cardiovascular diseases. Curr. Issues Mol. Biol. 22: 41–64. [DOI] [PubMed] [Google Scholar]
- 8.Chawla A., Boisvert W. A., Lee C. H., Laffitte B. A., Barak Y., Joseph S. B., Liao D., Nagy L., Edwards P. A., Curtiss L. K., et al. . 2001. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell. 7: 161–171. [DOI] [PubMed] [Google Scholar]
- 9.Oram J. F., Lawn R. M., Garvin M. R., and Wade D. P.. 2000. ABCA1 is the cAMP-inducible apolipoprotein receptor that mediates cholesterol secretion from macrophages. J. Biol. Chem. 275: 34508–34511. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y. W., Ma X., Li X. X., Liu X. H., Xiao J., Mo Z. C., Xiang J., Liao D. F., and Tang C. K.. 2009. Eicosapentaenoic acid reduces ABCA1 serine phosphorylation and impairs ABCA1-dependent cholesterol efflux through cyclic AMP/protein kinase A signaling pathway in THP-1 macrophage-derived foam cells. Atherosclerosis. 204: e35–e43. [DOI] [PubMed] [Google Scholar]
- 11.Gordts S. C., Singh N., Muthuramu I., and De Geest B.. 2014. Pleiotropic effects of HDL: towards new therapeutic areas for HDL-targeted interventions. Curr. Mol. Med. 14: 481–503. [DOI] [PubMed] [Google Scholar]
- 12.Levkau B. 2015. HDL-S1P: cardiovascular functions, disease-associated alterations, and therapeutic applications. Front. Pharmacol. 6: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sattler K., Graler M., Keul P., Weske S., Reimann C. M., Jindrova H., Kleinbongard P., Sabbadini R., Brocker-Preuss M., Erbel R., et al. . 2015. Defects of high-density lipoproteins in coronary artery disease caused by low sphingosine-1-phosphate content: correction by sphingosine-1-phosphate-loading. J. Am. Coll. Cardiol. 66: 1470–1485. [DOI] [PubMed] [Google Scholar]
- 14.Yanagida K., and Hla T.. 2017. Vascular and immunobiology of the circulatory sphingosine 1-phosphate gradient. Annu. Rev. Physiol. 79: 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swendeman S. L., Xiong Y., Cantalupo A., Yuan H., Burg N., Hisano Y., Cartier A., Liu C. H., Engelbrecht E., Blaho V., et al. . 2017. An engineered S1P chaperone attenuates hypertension and ischemic injury. Sci. Signal 10: eaal2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hla T., and Dannenberg A. J.. 2012. Sphingolipid signaling in metabolic disorders. Cell Metab. 16: 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi S., and Snider A. J.. 2015. Sphingolipids in high fat diet and obesity-related diseases. Mediators Inflamm. 2015: 520618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meikle P. J., and Summers S. A.. 2017. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat. Rev. Endocrinol. 13: 79–91. [DOI] [PubMed] [Google Scholar]
- 19.García-Arribas A. B., Alonso A., and Goñi F. M.. 2016. Cholesterol interactions with ceramide and sphingomyelin. Chem. Phys. Lipids. 199: 26–34. [DOI] [PubMed] [Google Scholar]
- 20.Róg T., and Vattulainen I.. 2014. Cholesterol, sphingolipids, and glycolipids: what do we know about their role in raft-like membranes? Chem. Phys. Lipids. 184: 82–104. [DOI] [PubMed] [Google Scholar]
- 21.Torres S., Balboa E., Zanlungo S., Enrich C., Garcia-Ruiz C., and Fernandez-Checa J. C.. 2017. Lysosomal and mitochondrial liaisons in Niemann-Pick disease. Front. Physiol. 8: 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worgall T. S. 2011. Sphingolipid synthetic pathways are major regulators of lipid homeostasis. Adv. Exp. Med. Biol. 721: 139–148. [DOI] [PubMed] [Google Scholar]
- 23.Pitman M. R., Pham D. H., and Pitson S. M.. 2012. Isoform-selective assays for sphingosine kinase activity. Methods Mol. Biol. 874: 21–31. [DOI] [PubMed] [Google Scholar]
- 24.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 25.Beljanski V., Knaak C., and Smith C. D.. 2010. A novel sphingosine kinase inhibitor induces autophagy in tumor cells. J. Pharmacol. Exp. Ther. 333: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao P., Peterson Y. K., Smith R. A., and Smith C. D.. 2012. Characterization of isoenzyme-selective inhibitors of human sphingosine kinases. PLoS One. 7: e44543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.French K. J., Schrecengost R. S., Lee B. D., Zhuang Y., Smith S. N., Eberly J. L., Yun J. K., and Smith C. D.. 2003. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res. 63: 5962–5969. [PubMed] [Google Scholar]
- 28.Xiong Y., Lee H. J., Mariko B., Lu Y. C., Dannenberg A. J., Haka A. S., Maxfield F. R., Camerer E., Proia R. L., and Hla T.. 2013. Sphingosine kinases are not required for inflammatory responses in macrophages. J. Biol. Chem. 288: 32563–32573. [Erratum. 2016. J. Biol. Chem. 291: 11465.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witting S. R., Maiorano J. N., and Davidson W. S.. 2003. Ceramide enhances cholesterol efflux to apolipoprotein A-I by increasing the cell surface presence of ATP-binding cassette transporter A1. J. Biol. Chem. 278: 40121–40127. [DOI] [PubMed] [Google Scholar]
- 30.Spiegel S., and Milstien S.. 2011. The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 11: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q., Tsujita M., and Yokoyama S.. 1997. Selective down-regulation by protein kinase C inhibitors of apolipoprotein-mediated cellular cholesterol efflux in macrophages. Biochemistry. 36: 12045–12052. [DOI] [PubMed] [Google Scholar]
- 32.Yamauchi Y., Hayashi M., Abe-Dohmae S., and Yokoyama S.. 2003. Apolipoprotein A-I activates protein kinase C alpha signaling to phosphorylate and stabilize ATP binding cassette transporter A1 for the high density lipoprotein assembly. J. Biol. Chem. 278: 47890–47897. [DOI] [PubMed] [Google Scholar]
- 33.Keul P., van Borren M. M., Ghanem A., Muller F. U., Baartscheer A., Verkerk A. O., Stumpel F., Schulte J. S., Hamdani N., Linke W. A., et al. . 2016. Sphingosine-1-phosphate receptor 1 regulates cardiac function by modulating Ca2+ sensitivity and Na+/H+ exchange and mediates protection by ischemic preconditioning. J. Am. Heart Assoc. 5: e003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camerer E., Regard J. B., Cornelissen I., Srinivasan Y., Duong D. N., Palmer D., Pham T. H., Wong J. S., Pappu R., and Coughlin S. R.. 2009. Sphingosine-1-phosphate in the plasma compartment regulates basal and inflammation-induced vascular leak in mice. J. Clin. Invest. 119: 1871–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato K., Malchinkhuu E., Horiuchi Y., Mogi C., Tomura H., Tosaka M., Yoshimoto Y., Kuwabara A., and Okajima F.. 2007. Critical role of ABCA1 transporter in sphingosine 1-phosphate release from astrocytes. J. Neurochem. 103: 2610–2619. [DOI] [PubMed] [Google Scholar]
- 36.Liu X., Ren K., Suo R., Xiong S. L., Zhang Q. H., Mo Z. C., Tang Z. L., Jiang Y., Peng X. S., and Yi G. H.. 2016. ApoA-I induces S1P release from endothelial cells through ABCA1 and SR-BI in a positive feedback manner. J. Physiol. Biochem. 72: 657–667. [DOI] [PubMed] [Google Scholar]
- 37.Zhao G. J., Yin K., Fu Y. C., and Tang C. K.. 2012. The interaction of ApoA-I and ABCA1 triggers signal transduction pathways to mediate efflux of cellular lipids. Mol. Med. 18: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bektas M., Allende M. L., Lee B. G., Chen W., Amar M. J., Remaley A. T., Saba J. D., and Proia R. L.. 2010. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J. Biol. Chem. 285: 10880–10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahn B. H., Grossman J., Ansell B. J., Skaggs B. J., and McMahon M.. 2008. Altered lipoprotein metabolism in chronic inflammatory states: proinflammatory high-density lipoprotein and accelerated atherosclerosis in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Res. Ther. 10: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Annema W., Willemsen H. M., de Boer J. F., Dikkers A., van der Giet M., Nieuwland W., Muller Kobold A. C., van Pelt L. J., Slart R. H., van der Horst I. C., et al. . 2016. HDL function is impaired in acute myocardial infarction independent of plasma HDL cholesterol levels. J. Clin. Lipidol. 10: 1318–1328. [DOI] [PubMed] [Google Scholar]
- 41.Annema W., Dikkers A., de Boer J. F., van Greevenbroek M. M., van der Kallen C. J., Schalkwijk C. G., Stehouwer C. D., Dullaart R. P., and Tietge U. J.. 2016. Impaired HDL cholesterol efflux in metabolic syndrome is unrelated to glucose tolerance status: the CODAM study. Sci. Rep. 6: 27367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keul P., Polzin A., Kaiser K., Graler M., Dannenberg L., Daum G., Heusch G., and Levkau B.. 2019. Potent anti-inflammatory properties of HDL in vascular smooth muscle cells mediated by HDL-S1P and their impairment in coronary artery disease due to lower HDL-S1P: a new aspect of HDL dysfunction and its therapy. FASEB J. 33: 1482–1495. [DOI] [PubMed] [Google Scholar]