Abstract
Background/purpose
Porphyromonas gingivalis is a major causative agent of chronic periodontitis, whilst circumstances for acquisition of the bacterium remain to be elucidated. To examine prevalence of the bacterium harboring distinct fimA types in dental plaque of children, we established PCR procedures that are applicable for specimens with limited amounts. By this method, all six fimA types including type I and Ib were directly identified, and prevalence of fimA types and their frequency of guardian-child transmission in Japanese children were assessed.
Materials and methods
Genomic DNA was purified from dental plaque specimens of 132 periodontally healthy children (2–12 years old, 4.8 ± 0.2 years) and 19 mothers of resultant P. gingivalis-positive child subjects. PCR-based fimA genotyping was performed, and untypeable strains in the first PCR analysis were determined by a nested PCR.
Results
P. gingivalis was found in 15.2% of the subjects (2–10 years old, 5.1 ± 0.6 years), and the most prevalent types were I and IV (37.0% each), followed by Ib and III (11.1% each), and then II (7.4%). Seven (35.0%) of the 20 P. gingivalis-positive subjects had combined colonization of type I with other fimA types. In most cases, bacterial prevalence and fimA types in the children were distinct from those of their mothers, indicating that its maternal transmission was not significant.
Conclusion
These results suggest that colonization of non-disease-associated fimA types I and IV P. gingivalis to the oral cavity initiates from early childhood without showing any periodontal inflammation.
Keywords: child, colonization, dental plaque, fimA, genotyping, Porphyromonas gingivalis
Introduction
Porphyromonas gingivalis, a Gram-negative black-pigmented anaerobe, has been implicated as one of the major causative agents of adult chronic periodontitis.1, 2 This bacterium colonizes the gingival sulcus via fimbriae, which attach to the gingival epithelial cells, salivary component proteins, and other subgingival plaque microorganisms. A major constituent of fimbriae is fimbrillin (FimA), which is encoded by the fimA gene classified into six genotypes (I–V, Ib).3, 4, 5, 6 Although the fine biochemical properties of fimbriae in each FimA type remain to be characterized, it has been reasonably suggested that fimbrillin types are closely related to pathogenetic potentials of this bacterium. Actually, epidemiological studies have demonstrated a high prevalence of type II fimA P. gingivalis from regions of adult periodontitis, indicating that this type is disease-associated. On the contrary, P. gingivalis with type I fimA has been predominantly found in periodontally healthy subjects, and then it is considered as a less pathogenic or non-disease-associated strain.6, 7, 8, 9, 10 It has not been clarified, however, what kinds of milieu or circumstances induce colonization of the pathogenic bacterial type. To address these issues, investigation of occurrence of P. gingivalis fimA types in the oral cavity of both healthy children and adults is worthwhile.
The fimA isoform genes are composed of 1,152–1,167 bp with a 43–85% amino acid sequence identity.11 Four of six fimA genotypes, i.e., II, III, IV, and V, can be simply identified by one-step PCR, while discrimination between type I and Ib requires an additional restriction test with RsaI6 or DNA sequencing12 of the PCR products, which makes fimA genotyping rather laborious. In addition, the previous data (0–37%)13, 14, 15, 16, 17 on P. gingivalis occurrence in healthy children are quite controversial, although colonization of the bacterium seems to initiate at the early stage of primary teeth eruption.13 These differences in the occurrence have been likely caused by limitations in the amounts of specimens obtained or the PCR procedures used for detection in previous studies.
In the present study, we established one-step PCR genotyping methods of all six P. gingivalis fimA types, which were combined with a nested PCR procedure to identify fimA types with untypeable specimens by the first PCR. Using these procedures, prevalence of P. gingivalis and their fimA types in periodontally healthy Japanese children were evaluated. In addition, the possibility of guardian-child transmission of the organism was assessed.
Materials and methods
A total of 132 children (71 males, 61 females, 2–12 years, 4.8 ± 0.2 years, mean ± S.D.), consulting the Department of Paediatric Dentistry of Iwate Medical University Dental Clinical Center, were selected for participation. The design of the study and procedures for obtaining informed consent were approved by the Ethics Committee of Iwate Medical University School of Dentistry (Approval no. D-01163). They were systemically healthy and showed no signs of gingival inflammation. The decay-filling-teeth (dft) rate was 16.7 ± 2.1%. Dental plaque specimens were collected from all primary teeth as well as subgingival locations when possible with sterilized instruments. To investigate bacterial transmission, subgingival dental plaque specimens were also obtained from the first molar of 19 mothers (35.2 ± 1.4 years) of the 20 P. gingivalis-positive children, including two sisters. Probing depths were <4 mm in all children and mothers, and none of the subjects had received professional dental cleaning or antibiotic medication in the three months prior to the examination. All plaque specimens were immediately suspended in 1 ml of sterilized phosphate buffered saline (pH 7.4), and then stored at −30 °C until use. As controls, subgingival plaque specimens were collected from four periodontally healthy adults not related to the children (3 males, 1 female; mean age = 29 years).
P. gingivalis ATCC 33277 (fimA type I, PGN_0180),18 16-1 (type Ib, AB058848),6 HW24D1 (type II, D17797),4 ATCC 49417 (type III, KF770043),19 W83 (type IV, AB795788),20 and HNA99 (type V, AB795792),12 as well as Streptococcus mutans ATCC 25175 (as a negative control) were grown as previously described.21, 22, 23 Genomic DNA was purified from clinical specimens and the bacterial cells using a genomic DNA purification kit (Promega, Madison, WI, USA), according to the manufacturer's instruction. The concentration of DNA dissolved in 10 mM Tris–HCl (pH 8.0) and 1 mM EDTA was determined by a Micro-Volume UV/Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and finally adjusted to 10 ng/μl. Total amount of DNA recovered from a specimen was 600 ng in average. DNA sample was divided into two or three tubes and stored at −80 °C until use.
The existence of bacterial DNA in the samples was initially confirmed by detection of the eubacterial 16S rRNA gene with DNA (1 ng) and a set of primers (Table 1) using AmpliTaq Gold 360 PCR Master Mix (Thermo Fisher Scientific). Subsequently, P. gingivalis prevalence was examined with a set of primers for the P. gingivalis 16S rRNA gene, and then fimA genotypes were investigated with a set of genotype-specific primers, including a novel set of primers to distinguish types I and Ib. The amplification cycles were as follows: 15 min at 95 °C for initial heat activation; and then a cycle for 15 s at 95 °C, 15 s at 58 °C, and 45 s at 72 °C. The number of PCR cycles was 30 for eubacterial and P. gingivalis 16S rRNA, fimA types II and V genes, and 40 for fimA types I, Ib, III, and IV genes.
Table 1.
Primers and PCR conditions used in this study.
| Target | Primer | Sequence (5′ – 3′) | Length (bp) | Ta (°C) | Elongation period (s) | Reference |
|---|---|---|---|---|---|---|
| 16S rRNAa | 16S-F | GGATTAGAGTTTGATCCTGGC | 728 | 55 | 40 | 24 |
| 16S-R | TACCTTGTTACGACTT | |||||
| Pg 16S rRNA | Pg 16S-F | TGTAGATGACTGATGGTGAAAACC | 198 | 58 | 15 | 25 |
| Pg 16S-R | ACGTCATCCCCACCTTCCTC | |||||
| fimAb | fimA uni-F | AAGTTTTTCTTGTTGGGACTTGC | 1,219 | 58 | 45 | This study |
| fimA uni-R | AACCCCGCTCCCTGTATTCCGA | |||||
|
fimA type Ic |
fimA I&Ib-F | CTGTGTGTTTATGGCAAACTTC | 173 | 58 | 15 | This study |
| fimA I-R | TTATTCTTAGGCGTATAATTGC | |||||
|
fimA type Ibd |
fimA I&Ib-F | CTGTGTGTTTATGGCAAACTTC | 173 | 58 | 15 | This study |
| fimA Ib-R | TTATTCTTAGGCGTATAACCAT | |||||
|
fimA type II |
fimA II-F | GCATGATGGTACTCCTTTGA | 292 | 58 | 15 | 26 |
| fimA II-R | CTGACCAACGAGAACCCACT | |||||
|
fimA type III |
fimA III-F | ATTACACCTACACAGGTGAGGC | 253 | 58 | 15 | 25 |
| fimA uni-R | AACCCCGCTCCCTGTATTCCGA | |||||
|
fimA type IV |
fimA IV-F | CTATTCAGGTGCTATTACCCAA | 249 | 58 | 15 | 25 |
| fimA uni-R | AACCCCGCTCCCTGTATTCCGA | |||||
|
fimA type Ve |
fimA V-F | TGGAACGAATACGCCTGAAGGA | 166 | 60 | 15 | This study |
| fimA uni-R | AACCCCGCTCCCTGTATTCCGA |
When a fimA genotype was not identified by the first PCR despite of positive for the P. gingivalis 16S rRNA gene, the two-step PCR procedure (nested PCR) was applied: At first, a DNA fragment was amplified with a set of fimA universal primers (fimA uni-F and fimA uni-R corresponding to P. gingivalis W83 genome 2237849–2237871 and 2239047–2239068, respectively) designed for common sequences among the six fimA genes using 40 cycles. Secondly, after purification of the products with Nucleo-spin gel and a PCR clean-up kit (Takara Bio, Kusatsu, Japan), the fimA-specific PCR was performed with an aliquot (1 μl) of the eluent containing the DNA fragment. Resultant PCR products were subjected to agarose gel electrophoresis using a 0.9% gel, and DNA stained with ethidium bromide was detected at 302 nm by use of ChemiDoc XRS Plus (Bio-Rad) Super Cool CCD Camera (resolution 1,392 × 1,040 pixels). PCR analysis for the P. gingivalis 16S rRNA gene and fimA genotyping was performed at least three times with each sample.
Results
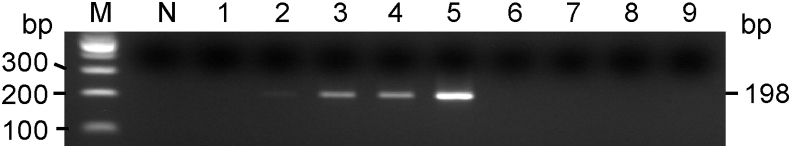
The specificity and sensitivity of PCR targeting P. gingivalis 16S rRNA gene was evaluated using genomic DNA purified from six P. gingivalis strains harboring every fimA type gene and from four periodontally healthy adults as controls. DNA amplicons were observed with a detection limit of 1 pg of DNA corresponding to approximately 4,000 bacterial cells (Fig. 1, lane 2, representative data on type I), while no PCR product was demonstrated even with 10 ng of DNA samples from the controls (Fig. 1, lanes 6–9) or S. mutans (data not shown). Similarly, precise specificity and detection limits of 1 pg were demonstrated on the remaining five fimA types (data not shown).
Figure 1.
Sensitivity of PCR for the P. gingivalis-specific 16S rRNA gene. PCR was performed with a set of primers and genomic DNA from P. gingivalis ATCC 33277 Lane M: 100-bp ladder; lane N: without DNA; lanes 1 to 5: 0.1, 1, 10, 100, and 1,000 pg DNA, respectively; lanes 6 to 9; 10 ng of DNA specimens obtained from periodontally healthy subjects.
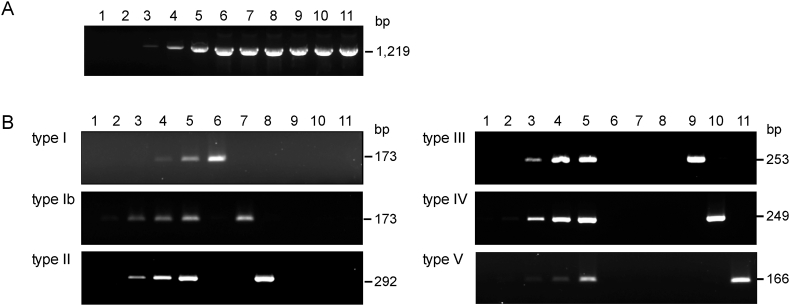
Since the discrimination between fimA types I and Ib by PCR has not been established yet, we newly designed PCR primer sets (Table 1). First, PCR with the fimA-universal primers provided amplicons in all six fimA types with a detection limit of 10 pg (Fig. 2A, representative data on type I). Secondly, the fimA type-specific PCR discriminated types I and Ib as well as the other types. This procedure provided detection limits of 10 pg of DNA from types Ib, II, III, IV and V, and 100 pg from type I (Fig. 2B). No amplification was observed even with 5 ng of DNA from non-corresponding fimA types. Hence, it was concluded that the present procedure was suitable to define all fimA genotypes. The variation in detection limits deemed to be inevitable due to the difference in annealing temperatures of PCR reactions, which were set to avoid cross amplification.
Figure 2.
Determination of fimA types by PCR. (A) PCR was performed with a set of fimA-universal primers. Lanes 1 to 5: 0.1, 1, 10, 100, and 1,000 pg DNA from P. gingivalis ATCC 33277, respectively; lanes 6 to 11: 5 ng DNA from ATCC 33277 (type I), 16-1 (type Ib), HW24D1 (type II), ATCC 49417 (type III), W83 (type IV), and HNA99 (type V), respectively. (B) PCR was performed with a set of fimA type-specific primers. Lanes 1–5: 0.1, 1, 10, 100, and 1,000 pg of corresponding DNA, respectively; lanes 6 to 11; 5 ng of DNA from types I, Ib, II, III, IV, and V, respectively.
Dental plaques were collected from 132 children (2–12 years old, mean age = 4.8 ± 0.2), and then genomic DNA was purified. The presence of bacterial DNA was confirmed by eubacterial 16S rRNA PCR in all samples, and the prevalence of P. gingivalis was found to be 20 of the 132 subjects (15.2%, mean age 5.1 ± 0.6 years). Among the P. gingivalis-positive specimens, fimA types were identified in 15 ones consisting of ten single and five multiple colonization (First PCR in Table 2) as follows; type IV was found in five subjects, type I and multiple colonization with type I and IV were detected in three each, and type II was detected in two subjects. Coexistence of type I with either Ib or III was found in two subjects.
Table 2.
fimA genotypes in the 20 P. gingivalis-positive children.
| fimA type | Detection n |
Total detection n (%) |
Prevalence n (%) |
||
|---|---|---|---|---|---|
| First PCR | Second PCRa | ||||
| Single | |||||
| I | 3 | 3 (15.0) | 10 (35.7) | ||
| Ib | 0 | 1 | 1 (5.0) | 3 (10.7) | |
| II | 2 | 2 (10.0) | 2 (7.2) | ||
| III | 0 | 1 | 1 (5.0) | 3 (10.7) | |
| IV | 5 | 1 | 6 (30.0) | 10 (35.7) | |
| V | 0 | 0 (0) | 0 (0) | ||
| Subtotal | 10 | 3 | 13 (65.0) | Total | 28 (100.0) |
| Multiple | |||||
| I, Ib | 1 | 1 | 2 (10.0) | ||
| I, III | 1 | 1 (5.0) | |||
| I, IV | 3 | 3 (15.0) | |||
| I, III, IV | 0 | 1 | 1 (5.0) | ||
| Subtotal | 5 | 2 | 7 (35.0) | ||
| Untypeable | 5 | 0 | 0 (0) | ||
The fimA genotypes of 20 P. gingivalis-positive children among the 132 investigated subjects were identified by one-step PCR (first PCR).
Five samples untypeable in the first PCR were further subjected to a nested PCR assay (second PCR).
Even on three trials, the fimA-type of the remaining five P. gingivalis-positive samples (25%) was not identified. Accordingly, for these samples, a two-step PCR procedure (nested PCR) was applied, and all fimA types were identified as three single and two multiple colonization (Second PCR in Table 2). To summarize the data of 20 P. gingivalis-positive subjects, single-strain colonization was found in 13 (65.0%) subjects and seven (35.0%) had multiple colonization, while fimA types I and IV were the most frequently detected (37.0% each), followed by Ib and III (11.1% each), and II (7.4%). None of the specimens had type V.
To investigate the possibility of guardian-child transmission of P. gingivalis, 19 mothers (35.2 ± 1.4 years) of the 20 P. gingivalis-positive children, including a sib pair, generously provided dental plaques for investigation. In the present cases, none of the guardians, who were all mothers taking their children to the hospital, showed symptoms of periodontal disease. Among them, 21.1% of the subjects (nos. 2, 3, 14, 15 in Table 3) were P. gingivalis-positive; P. gingivalis harboring fimA type II (n = 2), type IV (n = 1), and a combination of types I and II (n = 1) were observed. As for the bacterial transmission, occurrence of the common fimA types I and IV was observed in only two mother and child pairs (nos. 2 and 14), which suggests that maternal transmission of the organism is infrequent in the present subjects.
Table 3.
P. gingivalis fimA types of the child and mother subjects.a
| Child |
Mother |
|||
|---|---|---|---|---|
| Subject no. | Age (y) | Sex | fimA type | fimA type |
| 1 | 7 | M | I, III | ND |
| 2 | 5 | F | I, IV | IV |
| 3 | 5 | F | IV | II |
| 4 | 7 | M | II | ND |
| 5 | 3 | F | IV | ND |
| 6 | 9 | F | I, Ib | ND |
| 7 | 2 | F | I, III, IV | ND |
| 8 | 6 | F | IV | ND |
| 9 | 5 | F | IV | ND |
| 10 | 4 | F | IV | ND |
| 11 | 3 | M | II | ND |
| 12 | 10 | M | I | ND |
| 13 | 3 | M | III | |
| 14 | 4 | F | I | I, II |
| 15 | 9 | M | I | II |
| 16 | 2 | M | I, Ib | ND |
| 17 | 3 | M | IV | ND |
| 18 | 2 | F | I, IV | ND |
| 19 | 2 | M | Ib | ND |
| 20 | 10 | M | I, IV | ND |
Subjects no. 12 and 13 are a sib pair. fimA-types shown in italics were determined by nested PCR, while those in bold were identical in a mother and child pair. Four mothers were P. gingivalis-positive shown in shadow backgrounds. ND: not detected.
Transmission between mothers and children shows no statistical significance.
Discussion
This study presented the novel PCR procedure identifying all six P. gingivalis fimA genotypes in limited amounts of dental plaque specimens. fimA types I and Ib were successfully distinguished for the first time by one-step PCR. The newly designed primer-sets consisted of the common forward primer and two distinct reverse primers. The detection limits ranged from 10 to 100 pg of DNA, corresponding from 40,000 to 400,000 bacterial cells. By this procedure, the prevalence of P. gingivalis in healthy children was unambiguously determined as 15.2% (mean age = 5.1 ± 0.6 years old; range: 2–10 years), with no symptom of gingival inflammation. There was no statistical significance (p = 0.455, paired t test) between the mean age and gender between the P. gingivalis-positive (5.1 ± 0.6, 10/10) and those of the negative subjects (4.6 ± 0.2, 61/51). Of note, four subjects aged 2 years were positive, suggesting early colonization of this organism to the oral cavity. This figure of P. gingivalis prevalence was higher than those in previous examinations ranging from 0 to 6.3%.14, 15, 17 This difference may be caused by their limitations in the amounts of samples and the PCR procedures. Therefore, the present fimA-specific PCR procedure that includes sampling of dental plaque specimens, purification of genomic DNA, and PCR methods seems to provide accurate colonization profiles of P. gingivalis particularly in young children with primary teeth with poorly developed dental plaques.
Untypeable specimens have been also observed frequently (4–33%) in P. gingivalis-positive adults patients possibly because of altered detection limits of P. gingivalis and fimA PCR analyses.6, 27, 28 The present first PCR genotyping also resulted in five untypeable specimens (25.0%). To address their types, the two-step (nested) PCR was established. Consequently, fimA types I and IV were shown to be predominant (37.0% each), whereas the prevalence of type II was 7.4%. In addition, all cases of multiple colonization possessed type I fimA along with other fimA types. FimA types seem closely related to the bacterial pathogenicity. It was previously reported that periodontitis was associated with the occurrences of type I [odds ratio (OR) 0.16], type III (OR 1.96), type IV (OR 13.87), type V (OR 1.40), but much more significant with type II (OR 44.44).7 Thus, the present results indicated that non-disease-associated or lower pathogenic P. gingivalis fimA type I as well as type IV represents an earlier colonizer in the oral cavity. Similarly, predominant occurrence (50.7–100%) of fimA type I in periodontally healthy adults has been reported.6, 7, 8 Therefore, colonization of non-disease-associated P. gingivalis in the oral cavity may widely occur, leading no symptoms of periodontal inflammation. If this truly occurred, microbiological properties or pathogenicity might change among P. gingivalis strains harboring different fimbriae. We previously reported on activities of endo- and exo-peptidases that are considered to be its virulence factors in the strains. However, no significant relation of Lys- and Arg-gingipain activities and four kinds of dipeptidyl peptidase activities to fimA types was observed among the eight strains.22, 29 It is of interest to assess host circumstances and microbial factors that could be responsible for colonization of disease-associated P. gingivalis strains.
All P. gingivalis-positive child subjects were taken to the hospital by their mothers, and then the possibility of bacterial transmission from mother to child was examined. P. gingivalis was detected from child subjects aged 2–10 years, and thus the bacterium transmission from mother to child seemed to be likely. All the mother subjects were periodontally healthy (pocket depth < 4 mm) and the occurrence of P. gingivalis was 21.1% (4/19). This degree of prevalence was in approximate agreement with earlier studies of healthy adults, in which the prevalence was 16.8–23.1%.1, 8, 10 In addition, among four P. gingivalis-positive mother-child pairs, fimA type IV in subject no. 2 and fimA type I in no. 14 were coincidently detected, whereas subtypes of the other two pairs nos. 3 and 15 were different. Therefore, although the subject size was small and other family members had not been examined in this study, the present results suggest that P. gingivalis transmission scarcely occurred in our mother-child pairs, but it could be transferred from other family members.
Tuite-McDonnell et al.30 previously reported the prevalence of P. gingivalis and its vertical transmission among 564 members of 104 multigeneration families, in which the prevalence ranged from 50% in the great grandparents to 27.1% among children. Concordance in colonization was more frequently observed within entire families (p = 0.0000) and for spouses (p < 0.001), children and their mothers (p < 0.001), children and their fathers (p < 0.01). These data, therefore, indicated that P. gingivalis is commonly transmitted by contact with infected family members. Contrarily, in a smaller study with 78 subjects in 31 families, in which colonization was examined by culture, it was found that transmission of P. gingivalis between spouses was observed but its parent to child was limited.31 Since the higher prevalence (15.2%) of P. gingivalis in the child dental plaques was observed herein in contrast to previous figures (3.2–5.0%),15, 27, 31 we considered that P. gingivalis may be commonly acquired during childhood from colonized family members, and that it may also be transferred later in adults.
Vertical transmission of another periodontal bacterium, Actinobacillus actinomycetemcomitans, was estimated between 30% and 60%,32 and coincidence of the bacterium-positive parents and their children took place in 32% of family, suggesting more frequent vertical transmission than P. gingivalis.31, 32 These observations may suggest different environmental circumstances or the host oral homeostasis to allow the growth of P. gingivalis and A. actinomycetemcomitans. In addition, the cumulative meta-analysis demonstrated significant mother-to-child transmission of S. mutans, a pathogen of dental caries.33 This difference in transmitting routes may reflect the properties of S. mutans and P. gingivalis as initial and late colonizers, respectively.
In conclusion, this study is the first to establish a single-step PCR method for identification of fimA type I and Ib, and demonstrated that occurrence of P. gingivalis in Japanese children was 15% and non-disease-associated fimA types I and IV were predominant. These results indicate that colonization of this organism into the oral cavity may start in early childhood without showing any periodontal inflammation.
Conflicts of interest
The authors have no conflicts of interest relevant to this article.
Acknowledgments
The authors wish to thank all the children and their mothers for their valuable participation in this study. This study was supported in part by JSPS KAKENHI, Grant numbers 16K11481 (YON), 15K11047 (TKN), and 15K11020 (SK).
References
- 1.Yang H.W., Huang Y.F., Chou M.Y. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J Periodontol. 2004;75:1077–1083. doi: 10.1902/jop.2004.75.8.1077. [DOI] [PubMed] [Google Scholar]
- 2.Holt S.C., Ebersole J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 3.Dickinson D.P., Kubiniec M.A., Yoshimura F., Genco R.J. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J Bacteriol. 1988;170:1658–1665. doi: 10.1128/jb.170.4.1658-1665.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujiwara T., Morishima S., Takahashi I., Hamada S. Molecular cloning and sequencing of the fimbrilin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem Biophys Res Commun. 1993;197:241–247. doi: 10.1006/bbrc.1993.2467. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa I., Amano A., Kimura R.K., Nakamura T., Kawabata S., Hamada S. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J Clin Microbiol. 2000;38:1909–1914. doi: 10.1128/jcm.38.5.1909-1914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa I., Amano A., Ohara-Nemoto Y. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res. 2002;37:425–432. doi: 10.1034/j.1600-0765.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 7.Amano A., Kuboniwa M., Nakagawa I., Akiyama S., Morisaki I., Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79:1664–1668. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 8.Miura M., Hamachi T., Fujise O., Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodont Res. 2005;40:147–152. doi: 10.1111/j.1600-0765.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 9.Missailidis C.G., Umeda J.E., Ota-Tsuzuki C., Anzai D., Mayer M.P. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19:224–229. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao L., Wu Y.F., Meng S., Yang H., OuYang Y.L., Zhou X.D. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J Periodontal Res. 2007;42:511–517. doi: 10.1111/j.1600-0765.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 11.Hamada S., Fujiwara T., Morishima S. Molecular and immunological characterization of the fimbriae of Porphyromonas gingivalis. Microbiol Immunol. 1994;38:921–930. doi: 10.1111/j.1348-0421.1994.tb02148.x. [DOI] [PubMed] [Google Scholar]
- 12.Nagano K., Abiko Y., Yoshida Y., Yoshimura F. Genetic and antigenic analyses of Porphyromonas gingivalis FimA fimbriae. Mol Oral Microbiol. 2013;28:392–403. doi: 10.1111/omi.12032. [DOI] [PubMed] [Google Scholar]
- 13.McClellan D.L., Griffen A.L., Leys E.J. Age and prevalence of Porphyromonas gingivalis in children. J Clin Microbiol. 1996;34:2017–2019. doi: 10.1128/jcm.34.8.2017-2019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura S., Ooshima T., Takiguchi M. Periodontopathic bacterial infection in childhood. J Periodontol. 2002;73:20–26. doi: 10.1902/jop.2002.73.1.20. [DOI] [PubMed] [Google Scholar]
- 15.Ooshima T., Nishiyama N., Hou B. Occurrence of periodontal bacteria in healthy children: a 2-year longitudinal study. Commun Dent Oral Epidemiol. 2003;31:417–425. doi: 10.1046/j.1600-0528.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 16.Gafan G.P., Lucas V.S., Roberts G.J., Petrie A., Wilson M., Spratt D.A. Prevalence of periodontal pathogens in dental plaque of children. J Clin Microbiol. 2004;42:4141–4146. doi: 10.1128/JCM.42.9.4141-4146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai V.T., Campos M.R., Machado M.A.A.M., Lauris J.R.P., Greene A.S., Santos C.F. Prevalence of four putative periodontopathic bacteria in saliva of a group of Brazilian children with mixed dentition: 1-year longitudinal study. Int J Paediatr Dent. 2007;17:192–199. doi: 10.1111/j.1365-263X.2006.00813.x. [DOI] [PubMed] [Google Scholar]
- 18.Naito M., Hirakawa H., Yamashita A. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr J.E., Abramian J.R., Dao D.H. Genetic exchange of fimbrial alleles exemplifies the adaptive virulence strategy of Porphyromonas gingivalis. PLoS One. 2014;9:e91696. doi: 10.1371/journal.pone.0091696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson K.E., Fleischmann R.D., DeBoy R.T. Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda Y., Ohara-Nemoto Y., Kimura S., Ishibashi K., Kikuchi K. PCR-based identification of Staphylococcus epidermidis targeting gseA encoding the glutamic-acid-specific protease. Can J Microbiol. 2004;50:493–498. doi: 10.1139/w04-055. [DOI] [PubMed] [Google Scholar]
- 22.Nishimata H., Ohara-Nemoto Y., Baba T.T. Identification of dipeptidyl-peptidase (DPP)5 and DPP7 in Porphyromonas endodontalis, distinct from those in Porphyromonas gingivalis. PLoS One. 2014;9:e114221. doi: 10.1371/journal.pone.0114221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohara-Nemoto Y., Rouf S.M.A., Naito M. Identification and characterization of prokaryotic dipeptidyl-peptidase 5 from Porphyromonas gingivalis. J Biol Chem. 2014;289:5436–5448. doi: 10.1074/jbc.M113.527333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons S.R., Griffen A.L., Leys E.J. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38:2362–2365. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amano A., Nakagawa I., Kataoka K., Morisaki I., Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37:1426–1430. doi: 10.1128/jcm.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon J.H., Shin S.I., Chung J.H., Lee S.W., Amano A., Lee J.Y. Development and evaluation of new primers for PCR-based identification of type II fimA of Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2012;64:425–428. doi: 10.1111/j.1574-695X.2011.00889.x. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K., Nakano K., Nomura R. Distribution of Porphyromonas gingivalis fimA genotypes in Japanese children and adolescents. J Periodontol. 2005;76:674–679. doi: 10.1902/jop.2005.76.5.674. [DOI] [PubMed] [Google Scholar]
- 28.Beikler T., Peters U., Prajaneh S., Prior K., Ehmke B., Flemmig T.F. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003;111:390–394. doi: 10.1034/j.1600-0722.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 29.Nemoto T.K., Ohara-Nemoto Y. Exopeptidases and gingipains in Porphyromonas gingivalis as prerequisites for its amino acid metabolism. Jpn Dent Sci Rev. 2016;52:22–29. doi: 10.1016/j.jdsr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuite-McDonnell M., Griffen A.L., Moeschberger M.L., Dalton R.E., Fuerst P.A., Leys E.J. Concordance of Porphyromonas gingivalis colonization in families. J Clin Microbiol. 1997;35:455–461. doi: 10.1128/jcm.35.2.455-461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asikainen S., Chen C., Slots J. Likelihood of transmitting Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in families with periodontitis. Oral Microbiol Immunol. 1996;11:387–394. doi: 10.1111/j.1399-302x.1996.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 32.Van Winkelhoff A.J., Boutaga K. Transmission of periodontal bacteria and models of infection. J Clin Periodontol. 2005;(32 Suppl. 6):16–27. doi: 10.1111/j.1600-051X.2005.00805.x. [DOI] [PubMed] [Google Scholar]
- 33.da Silva Bastos Vde A., Freitas-Fernandes L.B., Fidalgo T.K. Mother-to-child transmission of Streptococcus mutans: a systematic review and meta-analysis. J Dent. 2015;43:181–191. doi: 10.1016/j.jdent.2014.12.001. [DOI] [PubMed] [Google Scholar]