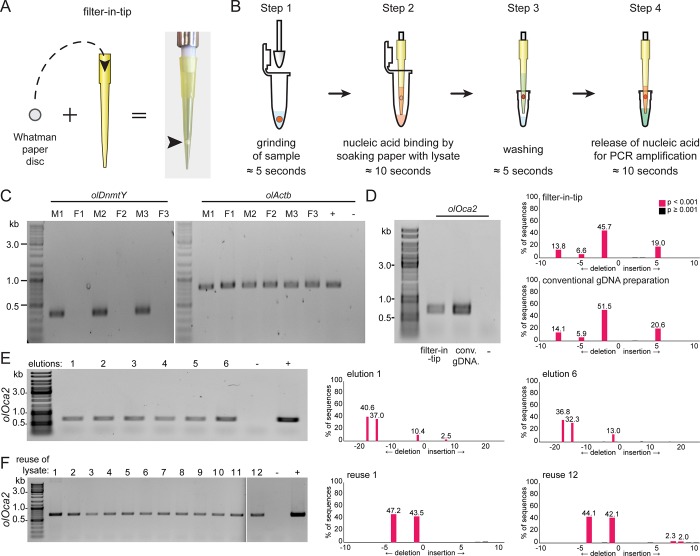
Fig 1. Filter-in-tips are highly sensitive for rapid transfer of nucleic acids for targeted genome editing diagnostics.
A) Assembly of filter-in-tip–Whatman filter paper / cellulose disc inserted into 200 μl yellow tip at the sample-proximal end. B) Schematic workflow of nucleic acid extraction using filter-in-tip. Step 1 lysis: pestle used to grind sample (red) in fin-clip buffer. Step 2 binding: using a filter-in-tip, pipet up lysate to let nucleic acids (red) bind to cellulose filter disc (soak for ≈10 sec). Release lysate back into tube for storage. Step 3 washing: wash filter disc containing nucleic acids (red) by pipetting nuclease free water in and out (≈5 sec). Step 4 elution: pipet up pre-mixed PCR mixture (wait ≈10 sec) to release nucleic acids and pipet back for amplification. C) Genomic male sex determination by olDnmtY PCR of 3 male (M1-3) and 3 female (F1-3) medaka fin clips. olActb amplification as controlNote: transfer of gDNA via filter-in-tip and amplification of loci was successful in all cases. D) Comparison between gDNA extracted with filter-in-tip (left) versus conventional gDNA preparation (right) of a single stage 32 medaka oca2 crispant. Note: matching allele distribution profiles of both amplicons (TIDE analysis). E) DNA retention capacity of a single filter-in-tip: cellulose filter soaked only once with a stage 32 medaka oca2 crispant lysate (10 seconds), washed once and eluted 6 individual times (10 seconds each). Note: matching allele distribution profiles of first and last amplicon (TIDE analysis). F) Repeated extraction of gDNA from a single stage 32 medaka oca2 crispant lysate using 12 filter-in-tips. Note: matching allele distribution profiles of first and last amplicon (TIDE analysis). wt gDNA (conventional gDNA preparation) as positive control (+), water as negative control (-).