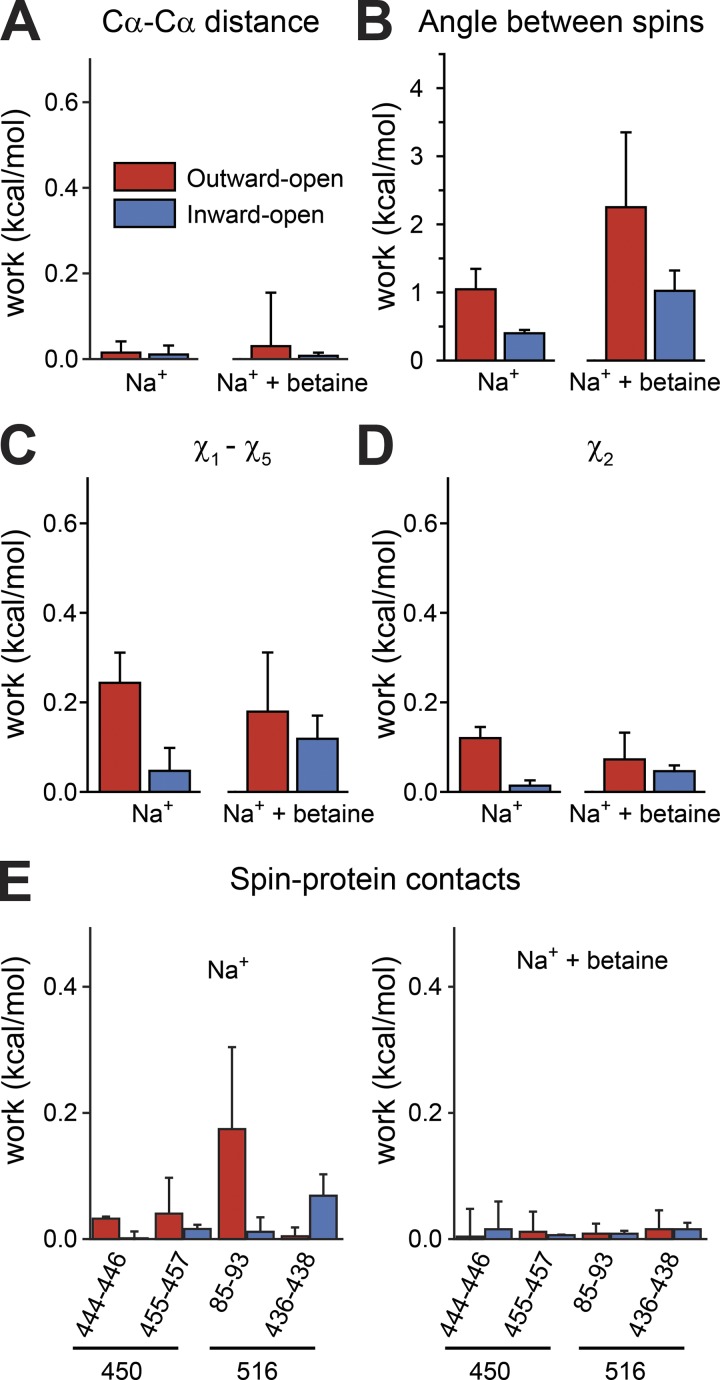
Figure 7.
Breakdown of contributions to the work for each molecular simulation system. Work computed with the Kullback-Leibler divergence between the biased and debiased trajectories (see Materials and methods) for the distance between the Cα atoms of residues 450 and 516 (A); the relative orientation of the spin labels (B); the side-chain dihedral angles χ1 to χ5 (C) or only χ2 (D); and interactions between the probes at positions 450 and 516 and nearby residues in loop EL5 (residues 436–438 or 444–446), TM8′ (455–457), or TM-1′ (85–93; E). Simulations were performed with two bound sodium ions (Na+), in either the absence (left) or presence (right) of betaine. The initial protein conformation was either outward- (PDB accession no. 4LLH chain A, red) or inward-facing (PDB accession no. 4C7R, blue).