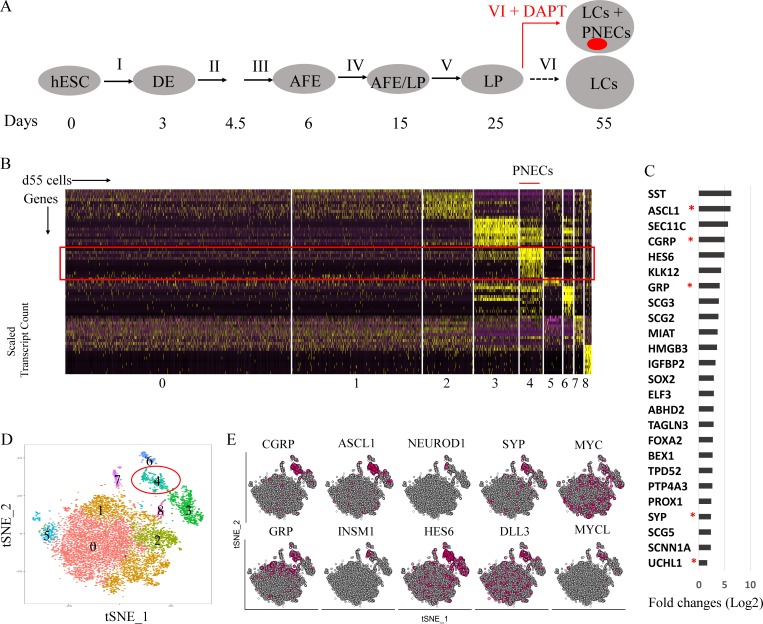
Figure 2.
Single-cell RNA profiling of RUES2-derived LCs in which NOTCH signaling was blocked by DAPT. (A) Schematic of the protocol used to generate PNECs by stepwise differentiation of hESCs and DAPT treatment from day 25 to 55. AFE, anterior foregut endoderm. (B–E) Single-cell RNA profiling of day 55 LCs derived from RUES2 cells treated with DAPT (5 µM) from day 25 to 55. (B) Heat map representing scaled expression of the most differentially expressed genes specific to different cell clusters. Rows represent genes, and columns represent cells. (C) Putative PNEC markers differentially expressed in the PNEC-like cell cluster number 4 in B. Bars indicate log fold change versus non-PNEC cells. Asterisks indicate canonical PNEC markers. (D) Reduced-dimensionality tSNE map colored by cluster assignment (Materials and methods section). (E) Individual cells positive for PNEC markers and other genes associated with neuroendocrine differentiation are denoted by red dots. Number of biological repeats (n) = 2.