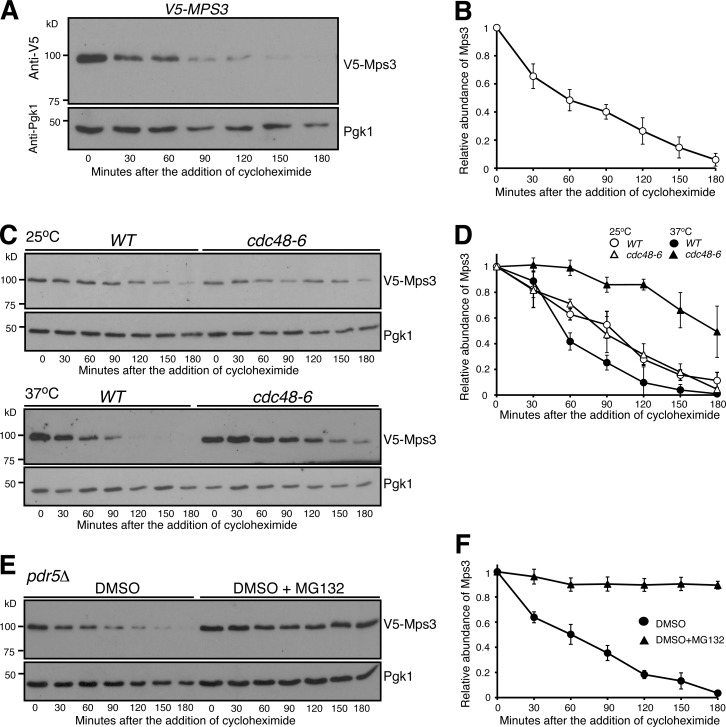
Figure 1.
Proteasome-dependent degradation of Mps3. (A and B) CHX-chase experiment showing Mps3 half-life. Yeast cells were grown to the exponential phase, then CHX was added to the culture media. Cell aliquots were withdrawn at indicated times, and protein extracts were prepared for Western blotting. An anti-V5 antibody was used to probe V5-Mps3. The level of Pgk1 serves as a loading control. Time zero is the point of CHX addition. Quantification of Mps3 protein abundance is shown in B. Error bars represent the standard deviation from the mean of seven biological replicates. (C and D) Cdc48 regulates Mps3 degradation. Cells were grown at 25°C to the exponential phase, and CHX was then added as shown in A. To inactivate cdc48-6, cells were shifted to 37°C for 1 h before the addition of CHX. Quantification of Mps3 protein abundance in shown in D. Samples were analyzed as in B; error bars represent the standard deviation from the mean of biological replicates (n = 3). Notably, the half-life of Mps3 at the nonpermissive temperature increased more than twofold. (E and F) The proteasome is responsible for Mps3 turnover. Protein extracts and Western blot were prepared as in A. The use of the pdr5Δ allele allows yeast to uptake MG132, which is dissolved in DMSO. Quantification of Mps3 protein abundance in shown in F. Samples were analyzed as in B. Error bars represent the standard deviation from the mean of biological replicates (n = 3). Note that Mps3 is stabilized in the presence of MG132.