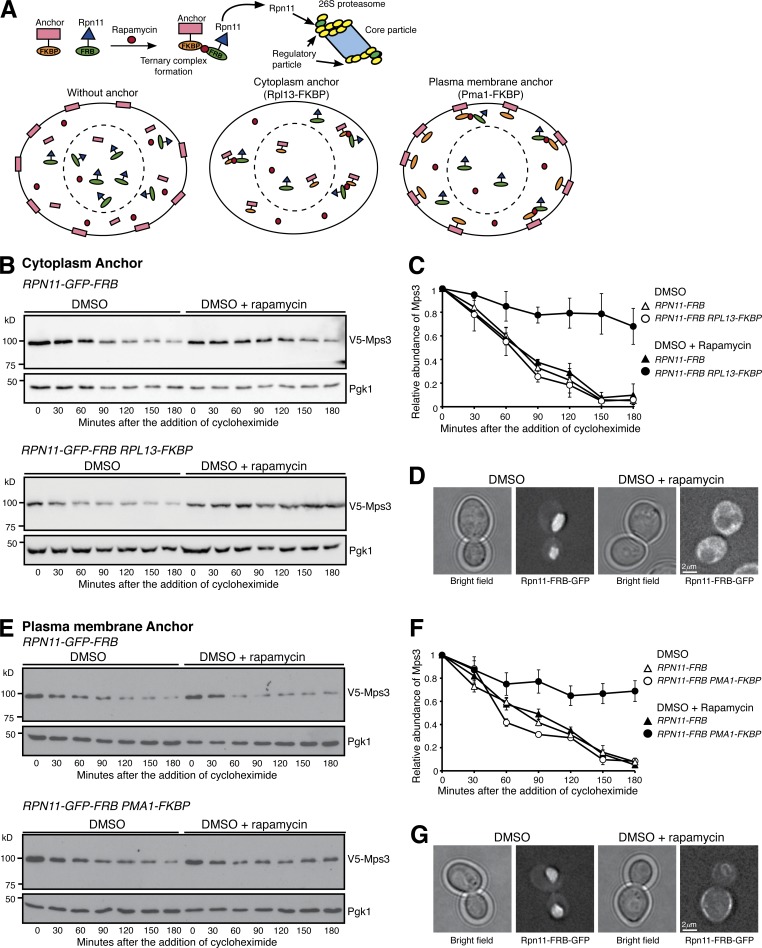
Figure 2.
Nucleus-localized proteasome regulates Mps3 degradation. (A) A schematic representation of forced proteasome localization using the anchor-away system. Briefly, Rpn11, a subunit of the 26S proteasome, was tagged with FRB, which in the presence of rapamycin induces a ternary complex formation between Rpn11-FRB and the FKBP-tagged anchor, either at the cytoplasm (Rpl13-FKBP) or at the plasma membrane (Pma1-FKBP). (B) Forced localization of proteasomes to the cytoplasm. Yeast cells were prepared for CHX chase, and rapamycin dissolved in DMSO was added to relocate the proteasome to the cytoplasm. (C) Quantification of Mps3 degradation as shown in B. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (D) Representative images showing forced localization of the proteasome to the cytoplasm 180 min after the addition of rapamycin. Note that Rpn11-GFP-FRB became dispersed in the cytoplasm in the presence of rapamycin. (E) Forced localization of the proteasome to the plasma membrane. Samples were analyzed as in B. (F) Quantification of Mps3 degradation as shown in E. Error bars represent the standard deviation from the mean of biological replicates (n = 3). (G) Representative images showing forced localization of the proteasome to the plasma membrane 180 min after the addition of rapamycin. Note that Rpn11-GFP-FRB was enriched at the cell periphery in the presence of rapamycin.