By performing an in vivo screen in Drosophila melanogaster, Scholz, Ehmann, et al. identify Complexin as a functional interaction partner of Bruchpilot. The two proteins mediate a physical attachment of synaptic vesicles to the active zone cytomatrix and promote rapid, sustained synaptic transmission.
Abstract
Information processing by the nervous system depends on neurotransmitter release from synaptic vesicles (SVs) at the presynaptic active zone. Molecular components of the cytomatrix at the active zone (CAZ) regulate the final stages of the SV cycle preceding exocytosis and thereby shape the efficacy and plasticity of synaptic transmission. Part of this regulation is reflected by a physical association of SVs with filamentous CAZ structures via largely unknown protein interactions. The very C-terminal region of Bruchpilot (Brp), a key component of the Drosophila melanogaster CAZ, participates in SV tethering. Here, we identify the conserved SNARE regulator Complexin (Cpx) in an in vivo screen for molecules that link the Brp C terminus to SVs. Brp and Cpx interact genetically and functionally. Both proteins promote SV recruitment to the Drosophila CAZ and counteract short-term synaptic depression. Analyzing SV tethering to active zone ribbons of cpx3 knockout mice supports an evolutionarily conserved role of Cpx upstream of SNARE complex assembly.
Introduction
In the nervous system, information is represented and processed by means of neuronal action potentials (APs). The ability of neurons to fire APs at high frequency places challenging demands on chemical synapses. To sustain the speed and temporal precision of synaptic transmission, presynaptic terminals must rapidly reload synaptic vesicles (SVs) at the active zone and prime them for exocytosis. During high-frequency stimulation, synapses often display short-term depression due to a transient drop in presynaptic neurotransmitter release. Many aspects of this phenomenon can be described by a limited pool of readily releasable vesicles (RRVs) at the active zone membrane, which is rapidly exhausted and then refilled from larger supply pools (Zucker and Regehr, 2002; Neher, 2015). The protein-rich cytomatrix at the active zone (CAZ) appears to play an important role in regulating such short-term synaptic plasticity by guiding SV replenishment (Zhai and Bellen, 2004; Südhof, 2012; Fernández-Busnadiego et al., 2013; Hallermann and Silver, 2013; Midorikawa and Sakaba, 2015). However, very little is known about the molecular mechanisms of SV reloading and the protein interactions that link SVs to the CAZ. This is because functional recordings of exo- and endocytosis provide only indirect information on processes preceding transmitter release, and low-affinity, transient interactions between SVs and the CAZ, which may be required for rapid vesicle fusion, can easily escape biochemical detection.
Bruchpilot (Brp) is an essential protein component of the Drosophila melanogaster CAZ (Kittel et al., 2006; Wagh et al., 2006). It shapes the filamentous CAZ structure by assembling as long polarized oligomers with its N terminus near Ca2+ channels at the active zone membrane and its C terminus extending into the cytoplasm (Fouquet et al., 2009; Ehmann et al., 2014). Functionally, Brp-dependent CAZ assembly is required for proper Ca2+ channel clustering to ensure adequate neurotransmitter release probability (pr; Kittel et al., 2006). Moreover, the very C-terminal region of Brp tethers SVs to the cytomatrix. At synapses of brpnude mutants, which lack the 17 C-terminal amino acids of Brp (∼1% of the protein), disrupted SV tethering is accompanied by short-term synaptic depression, impaired sustained transmitter release, and a slowed recovery phase (Hallermann et al., 2010b). Thus, Brp helps to establish release sites and accelerates the recruitment of SVs, enabling rapid and efficient excitation–secretion coupling at the active zone.
This basic understanding of Brp function provides an entry point to study molecular mechanisms of SV tethering to the CAZ and to shed light on protein interactions, which sustain ongoing synaptic transmission. Here, we devised an in vivo screen to search for vesicular interaction partners of Brp, including those with low affinity. Surprisingly, our results show that Complexin (Cpx), a key regulator of the core fusion machinery, participates in the SV cycle upstream of exocytosis. Besides interacting with the assembled trans-SNARE complex, this small, multifunctional protein also links SVs to Brp filaments and supports rapid SV recruitment to prevent short-term synaptic depression.
Results
Expression of Brp peptides in motoneurons alters SV localization
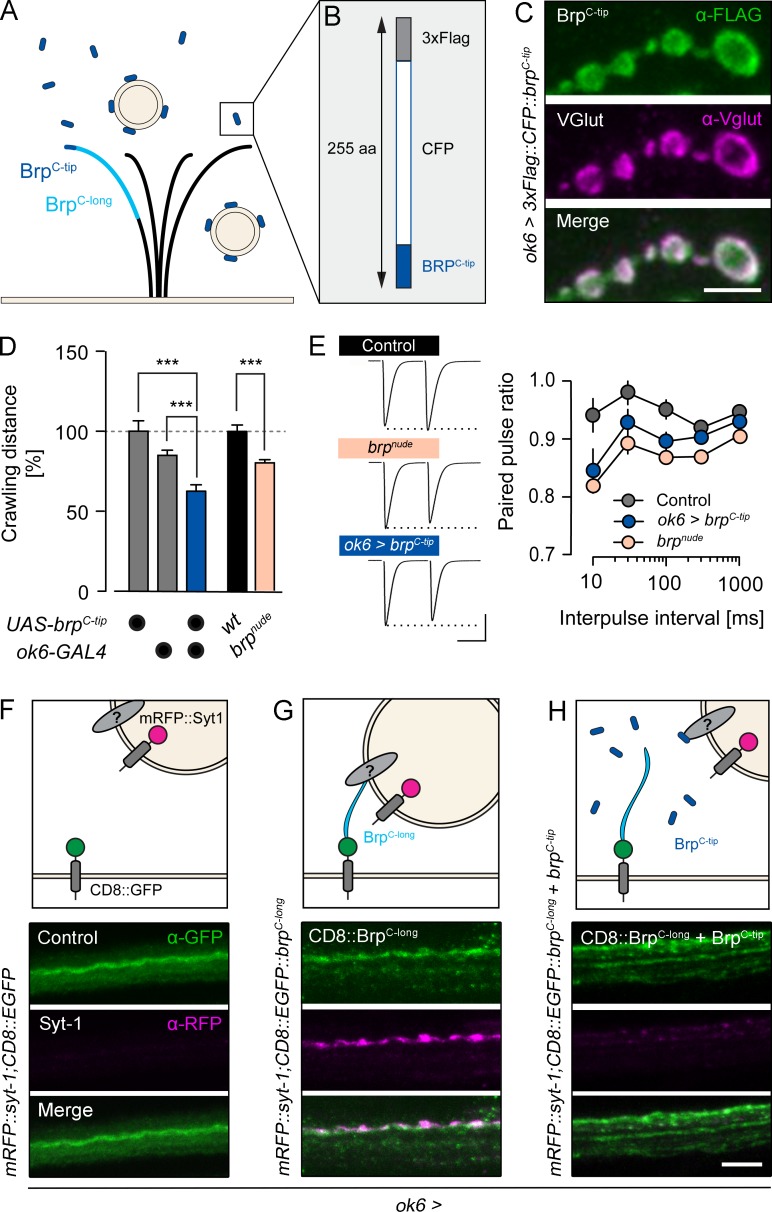
The 17 C-terminal amino acids of Brp (BrpC-tip hereafter) are required for efficient SV tethering to the CAZ (Hallermann et al., 2010b). We therefore tested whether a peptide encoding this amino acid sequence would in turn localize to SVs. To this end, we used the bipartite GAL4-UAS expression system (Brand and Perrimon, 1993) to drive a CFP and FLAG-tagged fusion construct of BrpC-tip in the cytoplasm of glutamatergic larval Drosophila motoneurons (Fig. 1, A and B; ok6>3xFlag::CFP::brpC-tip [brpC-tip]). As would be expected for an SV-bound peptide, immunostainings of the larval neuromuscular junction (NMJ) showed a clear signal overlap of BrpC-tip with the Drosophila vesicular glutamate transporter (VGlut; Fig. 1 C; Daniels et al., 2004).
Figure 1.
Neuronally expressed Brp peptides modify SV targeting. (A) Brp adopts a polarized orientation (light blue, approximately C-terminal half) to tether SVs near the active zone membrane. (B) A peptide containing the last 17 C-terminal amino acids of Brp (dark blue, BrpC-tip) fused to CFP and a FLAG-tag binds SVs. (C–E) Genetically expressed BrpC-tip (green, α-FLAG, ok6-GAL4 driver) colocalizes with SVs (magenta, α-VGlut) in the bouton cortex of motoneurons (C) and mimics the impaired locomotion (n ≥ 18; D), and paired-pulse depression of brpnude mutants (n = 12; E). (F–H) Top: Schematic illustrations of Brp-dependent SV enrichment in the axon. Bottom: Larval motor axons coexpressing mRFP::Syt-1 with CD8::EGFP (F), CD8::EGFP::BrpC-long, (G), and CD8::EGFP::BrpC-long + BrpC-tip (H). Maximal projections of confocal stacks stained against GFP (green) and RFP (magenta). Data are presented as mean ± SEM (Table S1). ***, P ≤ 0.001 (t test). Scale bars: (C) 3 µm; (E) 40 nA, 20 ms; and (F–H) 5 µm.
We reasoned that if Brp interaction partners exist on SVs, overexpressed cytoplasmic BrpC-tip would compete with endogenous, active zone resident Brp for the relevant vesicular binding sites. Functionally, this should phenocopy a C-terminal Brp truncation. brpnude larvae display impaired locomotion and pronounced short-term synaptic depression (Fig. 1, D and E; Hallermann et al., 2010b). Consistent with obstructed SV binding by Brp at the active zone, neuronal expression of BrpC-tip also decreased larval locomotion (Fig. 1 D, Fig. S1 A, and Table S1) and enhanced paired-pulse depression of AP-evoked excitatory postsynaptic currents (eEPSCs) at short interpulse intervals (Fig. 1 E and Table S2; rank sum test versus control: brpnude, 10 ms P = 0.0014, 30 ms P = 0.0086, all other intervals P ≤ 0.001; brpC-tip, 30 ms P = 0.0351, and 100 ms P = 0.0120; all other intervals NS). Collectively, these results show that BrpC-tip binds SVs.
Based on these findings, we investigated whether membrane-anchored transgenic Brp variants could be used to enrich SVs at ectopic sites. To localize Brp fragments to the axonal membrane distant from the NMJ, C-terminal segments of different length were fused to GFP-tagged CD8 (CD8::EGFP::brpC-x). For visualization, SVs were colabeled by Synaptotagmin-1 fused to an mRFP moiety (mRFP::syt-1), and both genetic constructs were expressed in motoneurons. Without the addition of a C-terminal Brp fragment, membrane-associated CD8::EGFP did not lead to the accumulation of SVs in motoneuron axons (Fig. 1 F). Including the short BrpC-tip sequence in the recombinant protein also failed to concentrate vesicles in the axon (Fig. S1 B). In contrast, when a longer fragment comprising roughly the C-terminal half of Brp (BrpC-long; Fig. 1 A) was fused to the CD8::EGFP carrier protein, SVs accumulated in the axon (Fig. 1 G). To control whether this effect is specifically mediated by membrane-anchored BrpC-long, we coexpressed cytoplasmic BrpC-tip and assayed ectopic SV localization. As expected, axonal enrichment of SVs was abolished (Fig. 1 H). This demonstrates that cytoplasmic and membrane-bound Brp fragments containing the C-terminal 17–amino acid peptide sequence compete with each other for SV targets (Fig. 1 H).
Reverse genetic screen for a Brp interaction partner
We made use of membrane-anchored BrpC-long and the concomitant SV clusters in the axon to screen for vesicular interaction partners of the Brp C terminus through RNAi. This strategy comprised scoring a selection of fly strains containing UAS-RNAi constructs, which interfere with mRNAs encoding presynaptic and vesicle-associated gene products (Table S3). Knockdown of molecules that mediate an interaction between Brp and the SV should disrupt complex formation by CD8::EGFP::BrpC-long and mRFP::Syt-1 at ectopic sites within the axon. This can be readily detected with light microscopy (Fig. 2 A). Importantly, screening takes place under physiological conditions, which should allow the capture of protein interactions with low affinity and specific molecular requirements in vivo. The screen also facilitates the identification of indirect interactors, i.e., molecules that mediate the link between Brp and the SV without necessarily being contacted by either molecule itself. Thus, we expected our screening protocol to exceed the sensitivity of standard biochemical approaches in its ability to identify functionally relevant molecular interactions at the CAZ–SV interface.
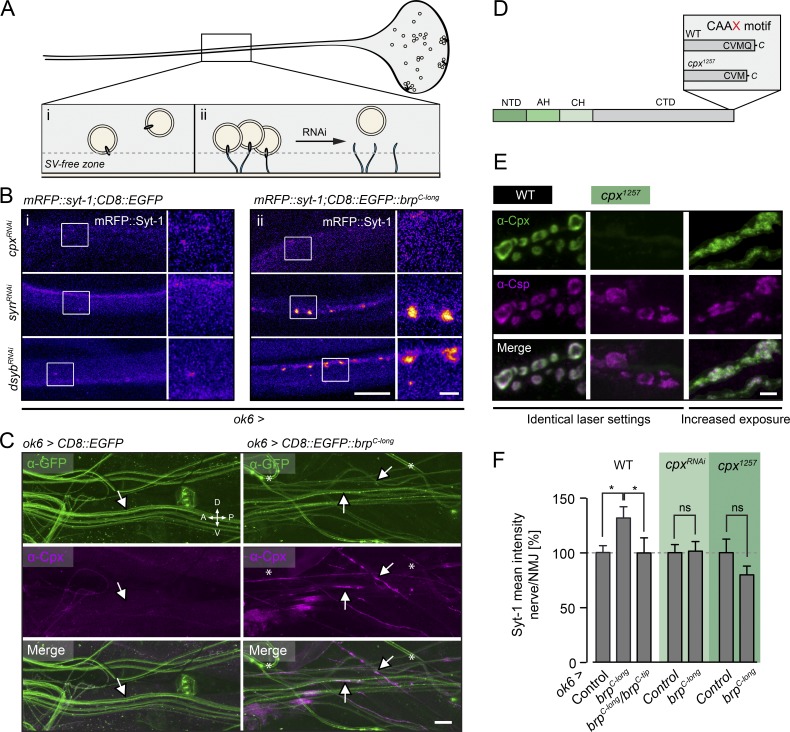
Figure 2.
Fluorescence-based genetic screen for tethering partners of Brp. (A) Screening strategy. The density of SVs is low in the motoneuron axon (i) and increases upon tethering to membrane-anchored Brp (ii; CD8::EGFP::BrpC-long). RNAi-mediated knockdown of factors mediating the SV–Brp interaction should prevent the axonal accumulation of SVs. (B) Confocal images of SVs (mRFP::Syt-1) in motoneuron axons in the absence (left, CD8::EGFP) and presence (right, CD8::EGFP::BrpC-long) of membrane-bound Brp (ok6-GAL4 driver). CpxRNAi prevents Brp-dependent localization of SVs in the axon, whereas other RNAi lines, e.g., synRNAi and dysbRNAi, have no discernible effect (Table S3). (C) Maximal projection of confocal images showing Cpx trapped in axons of ok6>CD8:EGFP::brpC-long (experimental condition), but not ok6>CD8:EGFP (control condition), larvae (arrows). Cpx was detected by a rb-α-Cpx antibody (Huntwork and Littleton, 2007), and GFP signals were enhanced using a ms-α-GFP antibody. Asterisks indicate unspecifically labeled tracheae. (D) Cpx layout, N-terminal domain (NTD), accessory helix (AH), central helix (CH), and C-terminal domain (CTD). Deletion of the terminal glutamine disrupts the CAAX motif in the cpx1257 mutant. (E) Maximal projections of confocal stacks show colocalization of Cpx (green, α-Cpx) with SVs (magenta, α-Csp) in WT motoneuron boutons (left). Cpx staining intensity is strongly reduced in cpx1257 boutons (center) and no longer matches the SV distribution (right; image taken with sixfold increase in laser power). (F) Quantification of nerve/NMJ ratio of the mRFP::Syt signal. The BRPC-long-dependent increase of SVs in the nerve is prevented by brpC-tip and cpxRNAi expression (ok6-GAL4) and in the cpx1257 mutant. Data are presented as mean ± SEM (n ≥ 13; Table S4). *, P ≤ 0.05 (rank sum test). Scale bars: (B) 10 µm, 2 µm (inset); (C) 30 µm; and (E) 3 µm. ns, not significant.
We tested 27 candidate genes (Table S3). Whereas knockdown of most targets did not noticeably perturb the BrpC-long–dependent enrichment of SVs in axons (examples shown for dysbindin [dysb] and synapsin [syn]; Fig. 2 B), motoneuronal expression of an RNAi construct directed against cpx (ok6>cpxRNAi) reduced axonal SV levels (Figs. 2 B and S1 C). Conversely, overexpression of CD8::EGFP::BrpC-long in motoneurons was accompanied by accumulation of Cpx in axons (Fig. 2 C). We therefore studied possible links among Cpx, SVs, and Brp in more detail.
The C terminus of the predominant Cpx isoform in Drosophila (Cpx7A) contains a membrane-binding farnesylation site (Fig. 2 D, CAAX motif; CVMQ; Zhang and Casey, 1996; Buhl et al., 2013). This motif is important for clamping spontaneous fusion events and for localizing Cpx to sites of exocytosis, possibly through an association with SVs (Buhl et al., 2013; Iyer et al., 2013). We therefore chose a cpx mutant, cpx1257, which lacks the last C-terminal glutamine and exhibits disrupted farnesylation (Iyer et al., 2013) to further investigate Cpx in the context of SV interactions (Fig. 2 D). At the WT NMJ, Cpx is distributed evenly around the bouton cortex, similar to vesicle proteins such as SV-associated Csp (cysteine string protein; Figs. 2 E and S2 B; Buhl et al., 2013). In contrast, cpx1257 mutants (cpx1257/cpxSh1) displayed strongly reduced Cpx levels at the NMJ (Figs. 2 E and S2 B). Here, the protein formed clusters primarily next to Brp profiles, indicating disturbed synaptic localization (Fig. S2 A; Buhl et al., 2013). Cpx enrichment at the cortex was drastically decreased, and the signal in interbouton regions and the main nerve fascicle appeared proportionally increased (Figs. 2 E and S2 B). In the WT ventral nerve chord (VNC), Cpx and Csp signals partially overlapped. In the cpx1257 mutant, however, Cpx was mainly concentrated in cell bodies (Fig. S2 C). Consistent with previous work (Buhl et al., 2013; Iyer et al., 2013; Robinson et al., 2018), these results suggest that C-terminal farnesylation of Drosophila Cpx promotes its association with SVs and its targeting to presynaptic terminals.
We quantified the contribution of Cpx to axonal SV targeting by BrpC-long. In the WT background, the ratio of SVs located in the nerve versus the NMJ was significantly increased upon expression of membrane-anchored BrpC-long and reverted by coexpression of cytoplasmic BrpC-tip (Fig. 2 F). Importantly, the nerve/NMJ ratio of SVs was leveled out in cpx1257 mutants and by cpxRNAi (Fig. 2 F and Table S4). These results indicate that Cpx supports SV tethering to Brp.
Cpx and Brp interact genetically
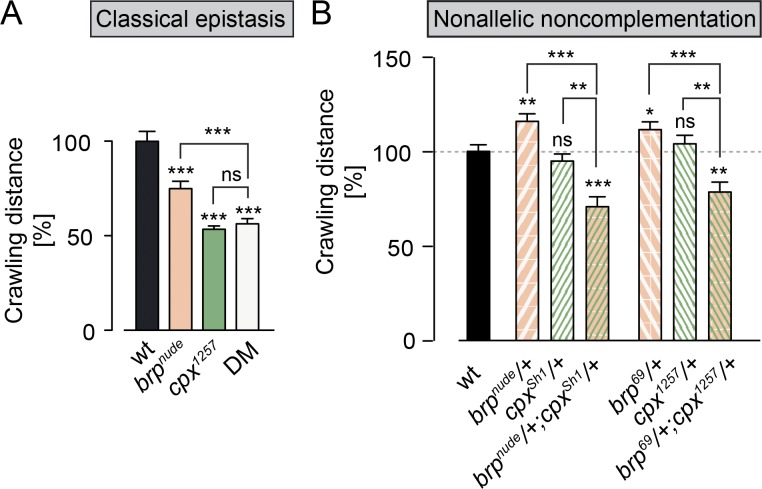
To substantiate these findings, we examined the genetic relationship between cpx and brp. Beginning with a classical epistasis assay, we tested locomotion of single (brpnude/brp69 or cpx1257/cpxSh1) and double mutant (DM; brpnude/brp69;cpx1257/cpxSh1) larvae. Both brpnude and cpx1257 animals displayed crawling defects. Notably, these phenotypes were not additive, implying that both proteins act in the same pathway at the synapse (Fig. 3 A and Table S1).
Figure 3.
Genetic interaction between cpx and brp. (A) Quantification of crawling distances covered by larvae over a period of 2 min. The severe locomotion defects of brpnude (brpnude/brp69) and cpx1257 (cpx1257/cpxSh1) single mutants are not additive in the DM (n ≥ 19, t test). (B) Whereas larvae carrying one mutant copy of cpx or brp crawl normally, the double heterozygotes (brpnude/+;cpxSh1/+ and brp69/+;cpx1257/+) display impaired locomotion (n ≥ 27, rank sum test). This supports the notion that Brp and Cpx function in a common signaling pathway. Data are presented as mean ± SEM (Table S1). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. ns, not significant.
Nonallelic noncomplementation describes the failure of recessive mutations in two distinct loci to complement one another. Put differently, the double heterozygote displays a phenotype despite the presence of a WT copy of each gene. This phenomenon often indicates a physical interaction between the two gene products. Moreover, nonallelic noncomplementation assays in Caenorhabditis elegans have demonstrated that synaptic function is sensitive to hypomorphic mutations, whose altered gene products may “poison” a limited number of presynaptic protein complexes (Yook et al., 2001). We therefore tested whether brpnude exhibited noncomplementation with a cpx-null allele (cpxSh1) and, vice versa, whether cpx1257 exhibited noncomplementation with a brp-null allele (brp69). Larval crawling measurements revealed impaired locomotion in the double heterozygotes, which were not observed in either heterozygote alone (Fig. 3 B and Table S1). These defects were synergistic and not additive effects of the noncomplementing mutations. Thus, brpnude and cpx1257 mutations sensitize synaptic function to decreased levels of Cpx and Brp, respectively. These results further support the view that Brp and Cpx share a functional pathway.
Disruption of Cpx targeting to SVs impairs their tethering to the CAZ
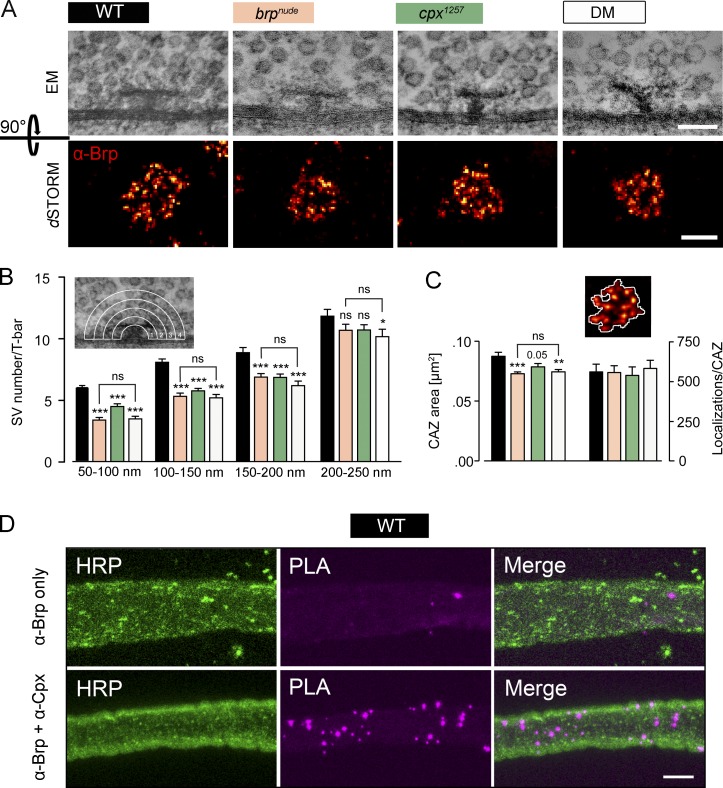
In EM, the Drosophila CAZ is frequently associated with an electron-dense, T-shaped protrusion (termed T-bar) extending inwards from the active zone membrane, with BrpC-tip marking its membrane-distal region (Fig. 4 A). As the brpnude mutant is characterized by a reduced number of SVs morphologically tethered to the T-bar (Hallermann et al., 2010b), we investigated whether the cpx1257 mutation also led to decreased SV tethering by the T-bar. Quantifying the number of SVs within four concentric shells from 50 to 250 nm around the T-bar (Fig. 4 B and Table S5) revealed that both brpnude and cpx1257 mutations impair SV tethering, leading to fewer SVs in the first three shells. Importantly, these morphological phenotypes were not additive in the DM. This strongly suggests that an interaction between Brp and Cpx helps tether SVs to the CAZ.
Figure 4.
Cpx supports SV tethering to the Drosophila CAZ. (A) Top: T-bars (viewed from the side) with associated SVs demark the CAZ in electron micrographs of larval motoneurons. Bottom: dSTORM images of antibody stainings against Brp (mAb nc82, NMJ 6/7) show the CAZ viewed en face. (B) Quantification of SVs surrounding the T-bar in four concentric shells, each 50 nm wide. Compared with the WT (black), fewer SVs are found near the T-bar in brpnude (beige) and cpx1257 (green) single mutants. A comparable, nonadditive effect is seen in the DM (white; n ≥ 38, rank sum test; Table S5). (C) The count of fluorophore localization events in dSTORM images is proportional to the number of Brp protein copies (Ehmann et al., 2014). In all three mutant genotypes, the same number of Brp proteins occupies a smaller area (n ≥ 18, t test; Table S6). This is in line with CAZ filaments spreading out upon SV tethering. (D) Maximal projections of confocal images showing that Brp and Cpx are located no further apart than ∼40 nm within WT larval axons using a PLA. The PLA was performed using primary antibodies against Brp and Cpx (lower row; PLA product, magenta) and its specificity was assessed by incubation with the Brp antibody only (upper row). Neuronal membranes were visualized with an antibody against HRP (green). Data are presented as mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (versus WT). Scale bars: (A) upper panel 100 nm, lower panel 200 nm; and (D) 5 µm. ns, not significant.
Using superresolution fluorescence imaging by direct stochastic optical reconstruction microscopy (dSTORM; Heilemann et al., 2008; van de Linde et al., 2011), it was recently demonstrated that the diminished tethering capacity of the brpnude CAZ is accompanied by subtle modifications of its nanoarchitecture (Ehmann et al., 2014). An average CAZ unit, equivalent to a T-bar, has been estimated to contain ∼137 Brp proteins. Hence, antibody staining against this major CAZ component can provide detailed information on the CAZ ultrastructure when interrogated by dSTORM with a spatial resolution of ∼20 nm (Ehmann et al., 2014). Viewed en face (i.e., with the optical axis perpendicular to the active zone membrane), dSTORM depicts the CAZ as a ring-like organization of Brp clusters, likely reflecting individual T-bar filaments. In brpnude mutants a normal number of Brp proteins are distributed over a smaller area (Fig. 4, A and C; and Table S6; Ehmann et al., 2014). This observation is consistent with a spreading out of T-bar filaments in the SV-tethered state. On the whole, the cpx1257 mutant CAZ was normally structured but, interestingly, also appeared subtly compacted. This ultrastructural phenotype was not additive in the DM. Thus, these results are consistent with the EM data (compare Fig. 4 B with Fig. 4 C) and the model that Brp and Cpx act in one pathway to tether SVs to the CAZ.
To delineate the spatial constraints of the interaction between Brp and Cpx at the active zone in more detail, we performed a proximity ligation assay (PLA). PLA with α-Brp and α-Cpx antisera yielded specific signals in the larval nerve (Fig. 4 D) and at the NMJ (Fig. S2 D), indicating that both proteins reside within a distance of 40 nm or less to each other (Söderberg et al., 2006). Because coimmunoprecipitation affinity purification of Cpx with the immobilized BrpC-tip peptide (Fig. S3 A) and coimmunoprecipitation of Cpx and Brp from lysates of animals expressing BrpC-long failed (Fig. S3 B), we considered a transient and/or low-affinity interaction. To address this possibility, we devised a photoaffinity labeling approach and designed a biotinylated photoreactive BrpC-tip peptide, with which we aimed to capture the potentially labile Brp–Cpx interaction into a covalent complex readily codetectable by α-Cpx antibodies and streptavidin, respectively. However, this approach also revealed no clear evidence for the formation of a Brp–Cpx complex (Fig. S3 C). Taken together, our data therefore suggest that Brp and Cpx most likely interact in an indirect manner.
Disruption of Cpx targeting to SVs enhances short-term synaptic depression
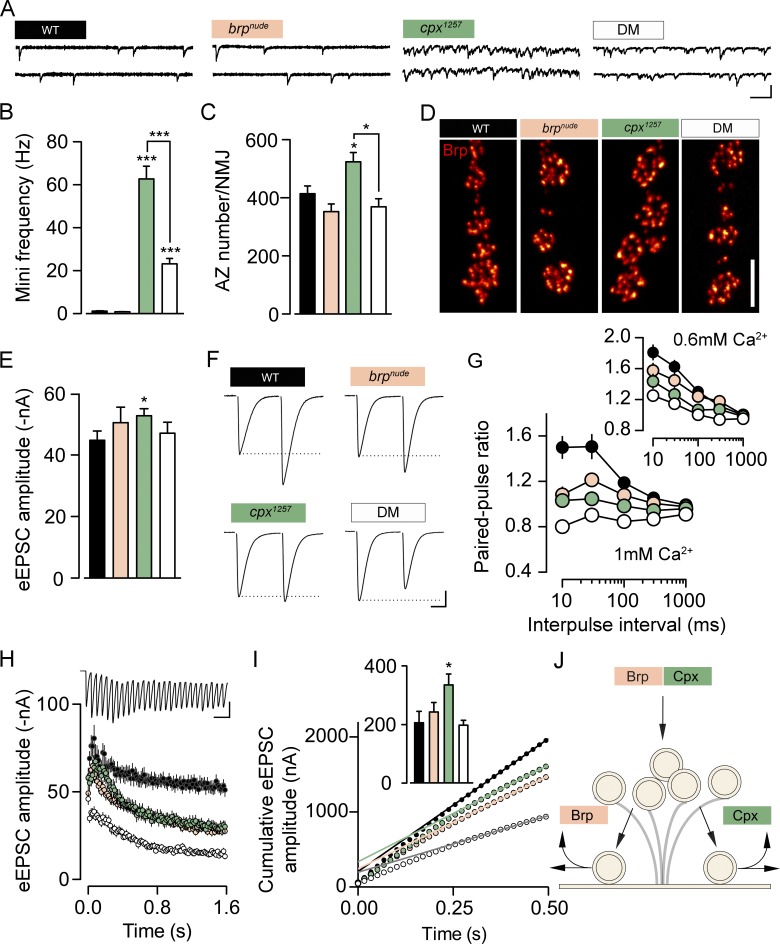
Next, we performed electrophysiological measurements to investigate synaptic function. As previously reported (Buhl et al., 2013; Iyer et al., 2013), C-terminal truncation of Cpx triggers a massive increase in the rate of spontaneous SV fusions, manifested as miniature excitatory postsynaptic currents (minis) in two-electrode voltage clamp (TEVC) recordings (Fig. 5, A and B; and Table S7). At the Drosophila NMJ, this effect appears to be caused by impaired clamping of the fusion machinery when Cpx is not localized properly to exocytosis sites. In cpx1257 mutants, the elevated mini frequency was accompanied by additional active zone formation (Fig. 5, C and D; and Table S8). This matches the NMJ outgrowth observed under conditions of increased spontaneous neurotransmitter release (Choi et al., 2014). The mini frequency of brpnude mutants was comparable to WT. Surprisingly, however, the brpnude allele partially rescued the high mini frequency of cpx1257 mutants and reverted the increased number of active zones (Fig. 5, A–D; and Tables S7 and S8). This highlights that Brp and Cpx perform distinct functions at the active zone. In contrast to Cpx, BrpC-tip is not critical for clamping SVs in a release-ready state and loss of BrpC-tip imposes a limit on the rate of spontaneous SV release, which occurs in the absence of the Cpx fusion clamp.
Figure 5.
Brp and Cpx accelerate SV reloading through separate pathways. (A and B) Example traces of TEVC recordings from larval NMJs (A) and quantification of data show the dramatically increased mini frequency of the cpx1257 mutant (green), which is partially rescued in the DM (B; white; n ≥ 14, rank sum test; Table S7). (C) Additional active zone formation accompanies the elevated mini frequency in the cpx1257 mutant (n ≥ 8, rank sum test; Table S8). (D) Shown are confocal images of Brp-stained NMJs (muscles 6/7). (E) The amplitude of eEPSCs is comparable in all genotypes at 0.2-Hz stimulation (n ≥ 10, t test; Table S7). (F and G) At short intervals, paired-pulse stimulation evokes less facilitation in the single mutants (brpnude, beige) than at WT synapses (black). This trend is further enhanced in the DM (n ≥ 10; Table S7). (H and I) Average eEPSC amplitudes at 60 Hz stimulation and cumulative amplitude plot (n ≥ 10; Tables S7 and S9). Back-extrapolation of a linear fit (0.3–0.5 s) yields an estimate of the RRV pool size (inset in nA; t test). (J) Schematic working model: Brp and Cpx cooperate to tether SVs to the CAZ and function in parallel pathways to support SV recruitment to the active zone membrane or release site clearance. Unless noted otherwise, TEVC recordings were made in 1 mM [Ca2+]e. Data are presented as mean ± SEM. *, P ≤ 0.05; ***, P ≤ 0.001. Scale bars: (A) 100 ms, 3 nA; (D) 5 µm; (F) 10 ms, 10 nA; and (H) 50 ms, 40 nA.
To obtain more information on synaptic physiology, we next analyzed eEPSCs. Despite normal basal current amplitudes at low frequency stimulation (0.2 Hz; Fig. 5 E and Table S7) both brpnude and cpx1257 synapses showed disrupted short-term facilitation during paired-pulse stimulation (Fig. 5, F and G; and Table S7; Hallermann et al., 2010b). Remarkably, this trend was further increased at DM synapses, resulting in low facilitation in 0.6 mM extracellular Ca2+ concentration ([Ca2+]e) and depression in 1 mM [Ca2+]e (Fig. 5, F and G; and Table S7; rank sum test versus control, 1 mM [Ca2+]e: brpnude, 10 ms P = 0.0039, 30 ms P = 0.0101, and 100 ms P = 0.02; all other intervals NS; cpx1257, 1,000 ms NS; all other intervals P ≤ 0.001; DM, 10 ms P = 0.0002, and 1,000 ms P = 0.0011; all other intervals P ≤ 0.0001; 0.6 mM [Ca2+]e: brpnude; all intervals NS; cpx1257, 10 ms P = 0.0123, 30 ms P = 0.0019, and 100 ms P = 0.0026; all other intervals NS; DM, 1,000 ms NS; all other intervals P ≤ 0.001). These observations point toward an additive effect of brpnude and cpx1257 mutations on short-term depression, which we investigated in more detail.
Desensitization of postsynaptic glutamate receptors shapes the decay time constant (τdecay) of eEPSCs and can contribute to use-dependent depression at the Drosophila NMJ (Heckmann and Dudel, 1997; DiAntonio et al., 1999; Schmid et al., 2008). Since the τdecay was not shortened in any of the mutant genotypes, enhanced receptor desensitization was likely not the cause of impaired facilitation (Fig. S4 A and Table S7). Presynaptic alterations that promote short-term depression without changing basal eEPSC amplitudes include slowed SV recruitment, as in the case of brpnude mutants, or an elevated pr of fewer RRVs (Ehmann et al., 2014). In both cases, normal SV release may be maintained during low-frequency stimulation, but as stimulation intervals shorten, SV availability becomes rate limiting and compound release drops. To differentiate between these two sources of depression, we estimated the size of the RRV pool in the different genotypes. Consistent with the paired-pulse protocols, a train of high-frequency stimulation (60 Hz) provoked similar short-term depression of eEPSC amplitudes at brpnude and cpx1257 synapses and triggered enhanced depression in the DM (Fig. 5 H and Table S7). Pool estimates can be derived by back extrapolation of a linear fit to the late phase of cumulatively plotted eEPSCs (Schneggenburger et al., 1999). While the quantitative interpretation of extrapolation-based pool estimates must be treated with caution (Hallermann et al., 2010a; Neher, 2015), we found no evidence of fewer RRVs in the single mutants or the DM (Fig. 5 I and Table S9). Thus, these results are consistent with slow SV reloading at brpnude and cpx1257 active zones. To obtain an independent mechanistic interpretation of SV release in the different genotypes, we employed an established modeling approach (Hallermann et al., 2010a; Weyhersmüller et al., 2011; Ehmann et al., 2014). To this end, a constrained short-term plasticity model was used to reproduce individual high-frequency trains and the recovery thereafter (Fig. S4, B–D). The simulations described impaired SV recruitment at both brpnude and cpx1257 active zones and, interestingly, also predicted a pr effect of the cpx1257 mutation. While such model predictions should not be overinterpreted, the sum of electrophysiological phenotypes in the DM indicates that Brp- and Cpx-dependent SV replenishment operate through separate mechanisms (Figs. 5 J and S4 D).
The altered short-term plasticity of cpx1257 mutants prompted us to interrogate the molecular organization of their active zones in more detail. Imaging by structured illumination microscopy (SIM; approximately twofold increase in 3D resolution compared with conventional widefield microscopy; Gustafsson et al., 2008) revealed an increased number of Ca2+ channel clusters at cpx1257 NMJs (Fig. S4, F and G; and Table S10) proportional to the elevated number of active zones (Fig. 5 C). Interestingly, whereas the arrangement of active zone Ca2+ channels is unaffected by the brpnude mutation (Hallermann et al., 2010b), the average cluster size is slightly reduced in cpx1257 mutants (Fig. S4, F and H; and Table S10). Future work will have to examine how this feature is related to SV release properties at cpx1257 active zones (Fig. S4 D).
Taken together, our results support a model whereby Brp and Cpx function in the same pathway to tether SVs to the CAZ (no additivity of ultrastructural phenotypes; Fig. 4) but act in parallel pathways to promote rapid recruitment of tethered SVs to the active zone membrane (additivity of short-term depression; Fig. 5 J).
SV tethering by Cpx is evolutionarily conserved
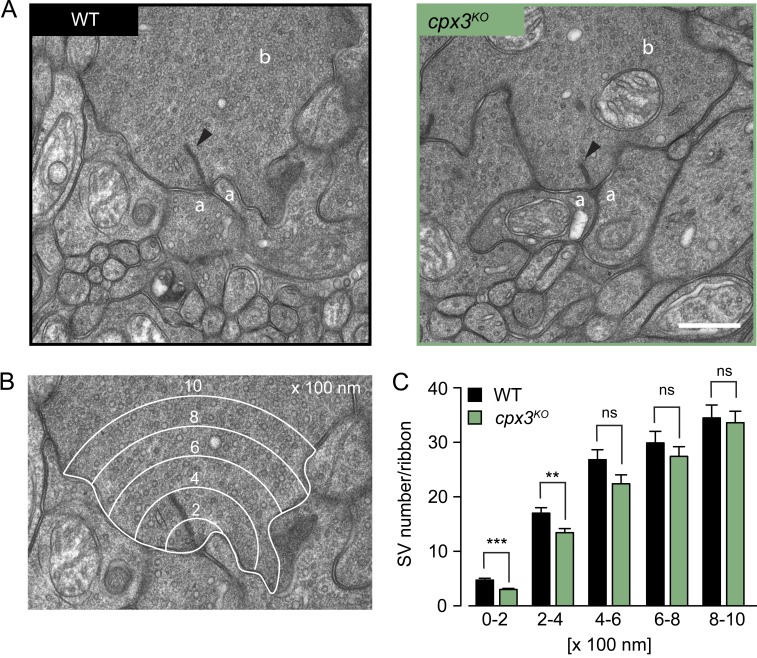
To test whether SV tethering is a specialization of Drosophila Cpx or an evolutionarily conserved property, we turned to rod bipolar cells (RBs) of the mouse retina. RB synapses onto AII amacrine cells are glutamatergic; they possess prominent electron-dense ribbons as CAZ specializations and express only Cpx3, a retina-specific isoform. Like the main isoform at the Drosophila NMJ, mammalian Cpx3 is also farnesylated at a C-terminal CAAX motif (Reim et al., 2005). We reanalyzed ultrastructural data from Mortensen et al. (2016) and focused specifically on the number of ribbon-proximal SVs in electron micrographs of RB terminals. In cpx3 knockout mutants (cpx3KO), significantly fewer SVs were found in concentric shells from 0 to 400 nm around the ribbon (Fig. 6 and Table S11), despite a normal cytoplasmic area covered by the shells (Fig. S5 and Table S11). This result resembles the morphological defect of cpx1257 mutants at the Drosophila NMJ and supports the notion that SV tethering is an evolutionarily conserved feature of certain Cpx family members.
Figure 6.
Mouse Cpx3 promotes SV tethering to ribbons of RBs. (A) Electron micrographs show WT (left) and cpx3KO (right) ribbon synapses of mouse RBs. The arrowheads indicate electron-dense ribbons to which SVs are tethered. Amacrine cell process (a), RB terminal (b). (B) Example of an electron micrograph overlaid with five concentric shells, each 200-nm wide (0–200 nm, 200–400 nm, 400–600 nm, 600–800 nm, and 800–1,000 nm), centered at the ribbon base. (C) Counting the SVs in each shell revealed a significant drop of SVs within 0–400 nm around cpx3KO ribbons. Data are presented as mean ± SEM (n ≥ 46; Table S11). **, P ≤ 0.01; ***, P ≤ 0.001 (rank sum test). Scale bar: 500 nm. ns, not significant.
Discussion
Tethering SVs to fine filamentous CAZ structures is believed to support their rapid reloading at the active zone membrane to sustain high neurotransmitter release rates (Hallermann and Silver, 2013). Correspondingly, disrupting CAZ tethers impedes ongoing SV release (Frank et al., 2010; Hallermann et al., 2010b,c; Snellman et al., 2011; Miskiewicz et al., 2014). The experiments reported here identify Cpx as a Brp interactor, which promotes SV tethering to the CAZ and counteracts short-term synaptic depression. Similarly, recent work on SV recycling at the Drosophila NMJ also found a reduced number of T-bar–associated SVs in cpxnull mutants (Sabeva et al., 2017). Cpx has been studied extensively in its capacity as a SNARE accessory protein, which regulates the fusion complex (Brose, 2008; Neher, 2010a; Trimbuch and Rosenmund, 2016; Reim, 2017). By interacting with the assembled trans-SNARE complex, Cpx promotes Ca2+-triggered evoked neurotransmitter release and, depending on species, synapse type, and preparation, either facilitates or clamps spontaneous release. At C. elegans and Drosophila NMJs, loss of Cpx leads to a dramatic increase in mini frequency (Huntwork and Littleton, 2007; Xue et al., 2009; Martin et al., 2011; Fig. 5, A and B; and Table S7), highlighting the bipolar function of Cpx at these invertebrate active zones. Here, Cpx appears to maintain SVs in a release-competent state by both blocking fusion and by supporting priming (Hobson et al., 2011). Our results indicate that the release-promoting role of Cpx extends even further upstream to facilitate SV recruitment to CAZ filaments and to the active zone membrane. This suggests that Cpx links the final stages of the SV cycle preceding exocytosis.
The hypomorphic cpx1257 allele strongly decreases Cpx protein levels and changes its localization pattern at the NMJ (Figs. 2 E and S2), consistent with previous reports linking C-terminal farnesylation of Cpx to synapse targeting (Buhl et al., 2013; Iyer et al., 2013; Robinson et al., 2018). Despite maintaining normal basal eEPSC amplitudes, the hypomorphic mutant displays a roughly 60-fold increase in mini frequency (Fig. 5, A and B; and Table S7). The different susceptibility of evoked and spontaneous fusion modes to reduced Cpx levels supports the notion that the clamping role of Cpx dominates at the Drosophila NMJ (Huntwork and Littleton, 2007; Xue et al., 2009; Iyer et al., 2013). Why does the C-terminal Brp truncation partially rescue the clamping defect of cpx1257 active zones? One explanation could be that this phenomenon is directly related to the additivity of short-term depression in the DM (Fig. 5 H and Table S7). If Cpx and Brp function in parallel to promote reloading of tethered SVs to the active zone membrane, then removing both pathways could limit both the maximal rate of evoked and (unclamped) spontaneous transmitter release. In brpnude single mutants, in turn, the intact Cpx clamp would prevent slowed reloading from reducing the mini frequency. Alternatively, evoked and spontaneous transmission may use nonidentical SV pools regulated differentially at the molecular level (Melom et al., 2013; Kaeser and Regehr, 2014; Crawford and Kavalali, 2015).
What could the parallel Cpx- and Brp-dependent pathways of SV replenishment correspond to mechanistically? It is tempting to speculate that Cpx may facilitate SV recruitment to activated release sites by helping to bring v-SNAREs and t-SNAREs within striking distance of each other to initiate fusion complex formation (Zenisek et al., 2000). On the other hand, Cpx may assist in release site activation itself (i.e., removal of SV proteins from prior fusion events and resetting of the release machinery). Given a limited number of release sites, their delayed restoration will enhance short-term depression (Kawasaki et al., 2000; Hosoi et al., 2009; Neher, 2010b). Similarly, Brp filaments may guide SVs to release sites or assist in site clearance. Therefore, an attractive model that describes parallel functional pathways for Brp and Cpx (producing additive short-term depression phenotypes in the DM) is that one of the two accelerates SV reloading and the other promotes release site activation. Extending this line of thought, we tested whether additional copies of Cpx could partially rescue the pronounced short-term depression of brpnude mutants (Fig. S4 E and Table S7). In this epistasis experiment, however, complementation did not occur. We therefore suspect that disrupting the joint tethering function of Cpx and Brp may create a bottleneck for ongoing transmitter release or that the number of functional Cpx interactors is limited.
Our morphological data point toward an interaction between Cpx and Brp in tethering SVs to the CAZ. Thus, defective tethering should contribute similarly to the functional phenotypes seen in both single mutants. SV tethering may enhance docking efficiency and rapid exocytosis by restricting the diffusion of SVs and thereby concentrating them in the vicinity of release sites. Consistent with this notion, findings at several synapses have suggested that undocked SVs located near the active zone membrane can be recruited and released very rapidly (Rizzoli and Betz, 2004; Jockusch et al., 2007; Hallermann et al., 2010b; Wang et al., 2016). Our results suggest that Cpx associates with SVs (Fig. 2 E and Fig. S2, A and B) and indicate that this interaction serves a dual function: it concentrates Cpx at synaptic sites and facilitates SV binding to Brp filaments. The association of Cpx with SVs has previously been reported in motoneurons of C. elegans, as well as larval and adult Drosophila. Whereas SV targeting and synaptic localization of Cpx depend on a C-terminal amphiphatic region in C. elegans, the predominant Drosophila isoform appears to associate with SVs through a membrane-binding farnesylated CAAX motif at its C terminus (Xue et al., 2009; Cho et al., 2010; Buhl et al., 2013; Iyer et al., 2013; Wragg et al., 2013). Mammalian Cpx isoforms 3 and 4 are also C-terminally farnesylated (unlike Cpx1 and 2), and here, too, this posttranslational modification is required for Cpx targeting to synapses (Reim et al., 2005, 2009). Drosophila Cpx is more mobile than SVs and less mobile than cytoplasmic GFP, indicating a low-affinity transient interaction with Cpx cycling on and off SVs (Buhl et al., 2013). Similarly, biochemically purified SVs from rat brain show that mammalian Cpx1 and 2 do not bind SVs with high affinity (Denker et al., 2011; Wilhelm et al., 2014), though the curvature-sensing C-terminal domain appears to preferentially localize Cpx1 to SV membranes (Gong et al., 2016). A transient, direct association between Cpx and Brp should have been detected via photoaffinity labeling in our biochemical analysis (Fig. S3 C). Hence, our in vivo setting was likely necessary to identify this indirect yet functionally relevant, protein interaction.
It is conceivable that our in vivo screen may have scored false negatives. That is, further SV proteins likely participate in CAZ tethering, and some of these may have passed undetected due to, for example, inefficient RNAi knockdown. Moreover, it should be noted that additional Brp domains and/or other CAZ proteins must be involved in tethering, since a physical association of SVs with the CAZ still persists in brpnude mutants, albeit at lower levels (Fig. 4 and Table S5; Hallermann et al., 2010b).
Of the four mammalian cpx genes, cpx1 and cpx2 are the main isoforms in the central nervous system. cpx3 and cpx4, in turn, are predominantly expressed in the retina, where they are specifically localized to ribbon synapses of photoreceptors and bipolar cells (Reim et al., 2005, 2009). Strikingly, knockout of cpx3 significantly reduced SV tethering to the RB ribbon (Fig. 6 and Table S11), highlighting intriguing parallels between this synapse and the Drosophila NMJ. In both glutamatergic neurons, C-terminal farnesylation of Cpx is required for its synaptic localization and at both active zones, Cpx participates in SV tethering to prominent electron-dense specializations of the CAZ (T-bar and ribbon at the NMJ and RB, respectively). This could indicate a specialized tethering function of farnesylated Cpx at ribbon-like CAZ structures. Perhaps relatedly, cpx3/4 double knockout abolishes the light-adapted reduction of SVs at the ribbon base of mouse photoreceptors (Babai et al., 2016). Although loss of Cpx in mouse hippocampal neurons did not cause any changes in the SV distribution at presynaptic active zones (Imig et al., 2014), we cannot exclude the possibility that Cpx may also have a role in SV tethering in other vertebrate central nervous system synapses. Either way, our findings argue that Cpx-dependent SV tethering is an evolutionarily conserved property of this multifunctional regulator of active zone function.
Materials and methods
Drosophila culture and stocks
Drosophila were cultured on standard cornmeal food at 25°C. The following strains were generated in this study: TAG61, w1118; P{UAS-3xFlag::amCFP::brpC17 w+}/CyO;; (BRPC-tip); TAG126, w1118; P{UAS-3xFlag::amCFP w+}/Cyo;; (CFP); TAG77, w1118;; P{UAS-mCD8::EGFP w+}attP2/TM3, Sb; (CD8::EGFP = no BRP); TAG70, w1118;; P{UAS-mCD8::EGFP::brpC17 w+}attP2/TM3, Sb; (CD8::EGFP::BRPC-tip); TAG118, w1118;; P{UAS-mCD8::EGFP::brpC862 w+}attP2/TM3, Sb; (CD8::EGFP::BRPC-long); TAG151, w1118; P{UAS-mRFP::syt-1 w+}attP40/Cyo;; (mRFP::SYT-1); and w*; P{UAS-cac1::EGFP} ok6-GAL4 w+/CyOGFP w−; cpxsh1/TM6b, Tb.
Additional strains are listed below. Flies were obtained from the Bloomington Stock Center (NIH P40OD018537), the Harvard Exelixis deficiency collection, or the Vienna Drosophila Research Center (RNAi strains; Dietzl et al., 2007) or were gifts from colleagues: w1118; brpnude/CyoactGFP w+;; (Hallermann et al., 2010b); w1118; brp69/CyoGFP w−;; (Kittel et al., 2006); w*;; cpxsh1/TM6b, Tb; (Huntwork and Littleton, 2007); w*;; cpx1257/TM6c (Iyer et al., 2013); w*; ok6-GAL4 w+;; (Sanyal, 2009); w*; Vglut-GAL4 w+;; (Daniels et al., 2008); w*;; Actin-5C-GAL4 w+; (Ito et al., 1997); w*; P{UAS-cac1::EGFP} ok6-GAL4 w+/CyOGFP w;; (Kawasaki et al., 2004); w1118;; Df(3R)Exel6140/TM6b, Tb; (BL# 7619; gift from F. Kawasaki, Pennsylvania State University, University Park, PA); w*;; P{UAS-EGFP::cpx}; (gift from F. Kawasaki); w*;; P{GD10482}v21477; (cpxRNAi); w*; P{GD9877}v33606;; (dunc13RNAi); w*;; P{GD8641}v43629; (tomosynRNAi); w*; P{GD14785}v29969;; (dCirlRNAi); w*; P{GD9502}v25291;; (dCAPSRNAi); w*; P{KK109431}v103201;; (cspRNAi); w*; P{GD1171}v47506;; (syt-12RNAi); w*;; P{GD8644}v24988; (syt-7RNAi); w*; P{GD15273}v39384;; (rim-1RNAi); w*;; P{GD12118}v27824/TM3; (rab3-GAPRNAi); w*;; P{GD11312}v26537/TM3; (gdiRNAi); w*; P{GD7330}v52438;; (rphRNAi); w1118 P{GD17411}v49247;;; (rac-1RNAi); w*;; P{GD10492}v34094; (rab5RNAi); w*; P{GD3785}v8784;; (sng-1RNAi); w*;; P{KK108941}VIE-260B; v110606 (synRNAi); w*;; P{GD11750}v27493 (annb9RNAi); w*;; P{GD14255}v36107; (annb10RNAi); w*;; P{GD10342}v25817; (twfRNAi); w*;; P{GD10754}v34354; (dysbRNAi); w*;; P{GD7149}v16133; (atg-1RNAi); w*; P{GD1254}v5702;; (rskRNAi); w*; P{GD12534}v35445;; (sap47RNAi); w*;; P{GD12383}v35346; (dsyd-1RNAi); w*; P{GD834}v2574;; (dvglutRNAi); w*; P{GD2233}v45726;; (vti1aRNAi); and w1118; P{GD2076}v10061/TM3; (mctpRNAi).
Transgene construction
pNH28 (BRPC-tip)
amCFP was amplified from pTL304 using nh_56F/57R and cloned into pENTRTM3C (catalog number A10464; Invitrogen) with KpnI and XhoI (pNH27). nh_57R contained the last C-terminal 17 amino acids of Brp. LR-based recombination of pNH27 with expression plasmid TFW provided the N-terminal 3xFlag and led to pNH28.
pNH54 (CFP)
Primers nh_56F/75R were used to amplify amCFP from pNH28. The resulting XhoI/KpnI-containing fragment was cloned into pENTRTM3C (pNH53). Subsequently, an LR reaction with pTFW resulted in pNH54.
pNH42 (no BRP)
The mCD8::EGFP fusion construct was amplified from pTL231 using primers tl_330F and nh_02R. The resulting 1.4-kb fragment, which carried a Kozak sequence, was cloned into pENTRTM3C using DraI and XhoI (pNH32). Next, the insert was recombined into pTW-attB using an LR recombination reaction, which yielded the expression construct pNH42.
pNH39 (BRPC-tip)
The mCD8::EGFP fusion construct was amplified from pTL231. Primers tl_330F and nh_3R were used to add a Kozak sequence, a 5x glycine linker, and a sequence encoding the last C-terminal 17 amino acids of Brp. Amplicon insertion into pENTRTM3C was done using DraI and XhoI and resulted in pNH33. LR recombination into pTW-attB led to the expression construct pNH39.
pNH52 (BRPC-long)
A 2.6 kb fragment encoding the last C-terminal 862 amino acids of Brp was amplified from pTL319 with primers nh_25F/24R. The amplicon was digested with DraI and XbaI and subsequently ligated with pNH43, which resulted in the ENTRY clone pNH51 and after recombination in the expression construct pNH52.
pNH60 (mRFP::SYT)
The amplification of syt from pTL143 was performed using primers nh_94F/95R. mRFP was amplified from pNH11 using nh_96F/97R. Both PCR fragments were cloned into pTW-attB using XhoI, NheI, and AgeI via triple ligation.
The AccuStar high-fidelity proofreading DNA polymerase (Eurogentec) was used for all PCR-based cloning steps. Initial insert verification was done by restriction analyses. To ensure the absence of errors, each PCR-amplified region was completely sequenced.
Primers (5′ to 3′ orientation)
See Table 1 for details on primer sequences.
Table 1. List of primers.
| Primer | Sequence |
|---|---|
| tl_330F | TACACTTTTAAATATCAACATGGCCTCACCGTTGACCCGCT |
| nh_02R | CCGCTCGAGCTACTTGTACAGCTCGTCCATGCCGAGA |
| nh_03R | CCGCTCGAGCTAGAAAAAGCTCTTCAAGAAGCCAGCTGGTCCAGCATCCTGCTGTTGCTGTTGCTGTCCGCCTCCGCCTCCGGGCTTGTAC |
| nh_24R | CTAGTCTAGACTAGAAAAAGCTCTTCAAGAAGCCAGCT |
| nh_25F | CCCAAGCTTGAGTTCGAAAAGATGCTGGAGAAGTAC |
| nh_56F | CGGGGTACCATCAACATGGCCCTGTCCAACAAGTTCATCG |
| nh_57R | CCGCTCGAGCTAGAAAAAGCTCTTCAAGAAGCCAGCTGGTCCAGCATCCTGCTGTTGCTGTTGCTGTCCGCCTCCGCCTCCGGGGAAGGGCACCACGGAGGTGATGTGG |
| nh_75R | CCGCTCGAGCTAGAAGGGCACCACGGAGGTGATGTGG |
| nh_94F | GCCCTCGAGATGCCGCCAAATGCAA |
| nh_95R | TCGGCTAGCTTACTTCATGTTCTTCAGGATCTCG |
| nh_96F | TAACCGGTCAACATGGCCTCCTCCGAGGACGTCATCA |
| nh_97R | CCGCTCGAGCGCAGCTGCAGCAGCGGCGCCGGTGGAGTGGCGGCCCTCG |
PhiC31-mediated integration of transgenes
Germline transformation of transgenes was done by BestGene Inc. either by random P-element transformation or site-specific phiC31 methodology (Groth et al., 2004). The vectors used were w+-marked allowing w+-based selection of recombinants. Specific integration sites attP40 (cyto site 25C6; Markstein et al., 2008) and attP2 (stock 8622; Bloomington Drosophila Stock Center; Groth et al., 2004) were used for transgene insertion on second and third chromosomes, respectively. Brp bait variants (CD8::EGFP, brpC-tip and brpC-long) were integrated into attP2 to ensure similar expression conditions.
Confocal microscopy
Wandering third-instar Drosophila larvae were dissected in ice-cold HL-3 (hemolymph-like solution; Stewart et al., 1994), fixed for 10 min using 4% paraformaldehyde in 0.1 M PBS and blocked for 30 min in PBT (PBS with 0.05% Triton X-100; Sigma-Aldrich) containing 5% normal goat serum (NGS; Jackson ImmunoResearch). Samples were incubated with primary antibodies at 4°C overnight. The next day, filets were incubated with secondary antibodies for 2 h at RT. Each antibody incubation step was followed by two short and three 20-min washing steps. Preparations were stored in Vectashield (Vector Laboratories) overnight at 4°C before mounting. Confocal recordings were performed at RT with a Zeiss LSM5 Pascal or Leica TCS SP8 confocal system. For Zeiss, microscopic datasets were collected with 63×, 1.25-NA or 100×, 1.4-NA objectives and photomultiplier tube (PMT) confocal detectors using the Zeiss ZEN software suite. For Leica, all samples were scanned through HC PL APO objectives (20×, 0.75 NA; 40×, 1.10 NA; 63×, 1.30 NA) immersed in 80% glycerol liquid (Leica type G, catalog number 11513910). Microscopic datasets were collected with PMT and HyD confocal detectors using the Leica LAS X SP8 software suite. To ascertain comparability of different genotypes, larvae were stained in the same vial and NMJs on muscles 6/7 (segments A2 and A3) were imaged in one session. Also, image acquisition of genotypes alternated and was performed with identical laser settings if not stated otherwise. Image analysis was done using ImageJ (National Institutes of Health). Prior to image analysis, unspecific signals were manually removed from maximal projections of confocal stacks. To analyze active zone numbers, the background was subtracted from the maximal projections. A threshold (mean gray value of 25) was set and a Gaussian blur (σ = 1 pixel) was applied to generate masks, which were subsequently superimposed onto the maximal projection. Active zones were detected with the find “Maxima” command and quantified via “Analyze Particles.”
To quantify SV levels, the mRFP::SYT signal was amplified using a polyclonal antiserum that recognizes RFP. For each larva, images were acquired at NMJ 6/7 (segment A2 or A3) and the nerve exiting the VNC at segment A8 (caudal end of the VNC). Laser settings were adjusted according to SV levels at the NMJ and subsequently retained to capture SV abundance in the nerve. This procedure was used to calculate the mRFP::SYT ratio of an individual larva and controlled for differences in expression strength across samples. After subtracting the background from maximal projections of confocal stacks, nonsynaptic signals were manually removed from NMJ images, a minimal threshold was set (individually adjusted for each sample), a Gaussian blur (σ = 1 pixel) was applied and the average intensity of mRFP::SYT was measured. In the nerve, the mean mRFP::SYT signal intensity was measured several times within an area of defined size (54.1 µm2). The brightest region was used to calculate the NMJ/nerve ratio of SVs.
Antibodies were used in the following dilutions: mouse α-Brp (nc82, 1:250; RRID: AB_528108; provided by E. Buchner, University Hospital Würzburg, Würzburg, Germany; Wagh et al., 2006), mouse-α-Csp (1:100; ab49, RRID: AB_2307345; Developmental Studies Hybridoma Bank), rabbit-α-Cpx (1:500; RRID: AB_2568068; Huntwork and Littleton, 2007), rabbit-α-VglutN-term (1:500; RRID: AB_2567386; provided by A. DiAntonio, Washington University School of Medicine, St. Louis, MO; Daniels et al., 2008), mouse-α-GFP (1:500; RRID: AB_259941; Sigma-Aldrich), rabbit-α-RFP (1:500; RRID:AB_10781500; Antibodies-Online), mouse-α-FLAG (1:500; RRID: AB_262044; Sigma-Aldrich), α-Hrp conjugated with Cy3 (1:250; RRID: AB 2338959; Jackson ImmunoResearch), Alexa Fluor 488–conjugated goat-α-mouse (RRID: AB2534069), goat-α-rabbit (RRID: AB_143165, both 1:250; Invitrogen Life Technologies), Cy3-conjugated goat-α-rabbit (RRID: AB_2338000), and goat-α-mouse antibodies (RRID: AB_2338690, both 1:250; Jackson ImmunoResearch).
PLA
The experiment was performed according to Wang et al. (2015) using the Duolink In Situ Red Kit Mouse/Rabbit (DUO92101; Sigma-Aldrich). Briefly, subsequent to standard dissection and fixation, L3 larvae were blocked for 1 h in PBT containing 5% NGS. Next, samples were incubated with rabbit-α-Cpx (1:500) and mouse-α-Brp (1:250) antibodies overnight at 4°C. After three 10–min washing steps using 0.05% PBT, the samples were incubated for 2 h at 37°C with 1:5 dilution of α-rabbit PLUS (DUO92002; Sigma-Aldrich) and α-mouse MINUS (DUO92004; Sigma-Aldrich) PLA probes, which were added to 5% NGS containing blocking solution. Alexa Fluor 488–conjugated α-HRP antibody (1:250) was added to the mixture in order to enable visualization of neuronal membranes. Following two 5-min washes with Wash buffer A, samples were treated with Ligation solution (1:40 dilution of Ligase in Ligase buffer) for 1 h at 37°C. Again, samples were washed twice for 2 min with Wash buffer A. Next, the samples were incubated in Amplification solution (1:80 dilution of polymerase in amplification buffer) for 2 h at 37°C. After two 10-min washing steps using Wash buffer B, samples were washed twice in 0.01× Wash buffer B and kept in Vectashield-H1000 overnight at 4°C before mounting.
Superresolution microscopy
SIM
In principle, sample preparation followed the procedures described for confocal imaging. Control and mutant larvae were stained in the same vial, mounted in Vectashield, and imaged in one session with the same laser settings to enable comparability. Images were acquired at 20°C from NMJs on muscle 4, segments A2 and A3, with a Zeiss Elyra S.1 equipped with a sCMOS camera (pco.edge 5.5 m), an oil-immersion objective (Plan-Apochromat 63×, 1.4 NA), and 488-nm and 641-nm lasers. The Z step size was set to 0.2 µm, and imaging was performed using five rotations of the grating at five different phase steps. Fourier transformation of SIM images was performed using ZEN software (Carl Zeiss), and subsequent quantifications were done with ImageJ. The background was subtracted from maximal projections and a gaussian blur (σ = 1) was applied to all images. A minimum threshold was applied depending on the epitope quantified thus generating a mask. A minimal overlay of this mask with the original image was created and subsequent particles were analyzed with the appropriate sizing criteria for Brp and Cacophony signals. Genotypes were blinded to the investigator.
The following antibodies were used: mouse-α-Brp (nc82, 1:100; RRID: AB_2314866; DSHB), GFP-Booster ATTO647N (1:100; alpaca; RRID: AB_2629215; ChromoTek), and Alexa Fluor 488–conjugated goat-α-mouse (1:250; RRID: AB_2534069; Thermo Fisher Scientific).
dSTORM
Superresolution imaging via dSTORM was performed as previously described (Ehmann et al., 2014). Mouse-α-Brp (nc82) was used at a concentration of 1:2,000 to quantify CAZ size and Brp localizations within the CAZ. Secondary goat-α-mouse F(ab′)2 fragments (A10534; Invitrogen) were labeled with Cy5-NHS (PA15101; GE Healthcare), yielding a degree of labeling of 1–1.3, and were used at a concentration of 5.2 × 10−8 M. Prior to dSTORM recordings, larval filets were embedded in photoswitching buffer, i.e., 100 mM mercaptoethylamine, pH 8.0, and enzymatic oxygen scavenger system (5% [wt/vol] glucose, 5 U/ml glucose oxidase, and 100 U/ml catalase; Schäfer et al., 2013), and arranged on an inverted microscope (Olympus IX-71) equipped with an oil- (60×, 1.49 NA) or water (63×, 1.15 NA)-immersion objective and a nosepiece stage (Olympus IX2-NPS; van de Linde et al., 2011). Positioning of filters and mirrors on a translational stage enabled switching between widefield and low-angle/highly inclined thin illumination (Sharonov and Hochstrasser, 2007; Tokunaga et al., 2008; van de Linde et al., 2011). Optical components for excitation and emission of Cy5 are described elsewhere (Ehmann et al., 2014; Paul et al., 2015). Final pixel size was 126 nm (oil-immersion objective) or 109 nm (water-immersion objective). Data were recorded at RT using an electron-multiplying charge-coupled device camera (iXon Ultra 897, Solis Software; Andor Technology). Each superresolved image consisted of 15,000 frames and was reconstructed using rapidSTORM (Wolter et al., 2010, 2012). A subpixel binning of 10 nm was applied, and fluorescence spots yielding >1,000 photons were included in the analysis. To quantify CAZ size and Brp localizations within the CAZ, masks were created (Gaussian blur σ = 1 pixel), minimally thresholded (0.15 counts), and overlaid with the original image. Individual CAZs from NMJ 6/7 (segments A2 and A3) were identified via their area (300 pixel infinity). Image analysis was performed using ImageJ. All genotypes were stained in the same vial and imaged in two sessions. To assure comparative imaging settings, data were analyzed for unspecific background labeling and excluded from the analysis if the background exceeded 2.3 single spots per square micrometer.
EM
Drosophila NMJ
Wandering third-instar larvae were dissected in ice-cold HL-3 (Stewart et al., 1994) and fixed with glutaraldehyde solution (2.5% glutaraldehyde, 50 mM cacodylate buffer, pH 7.2, 50 mM KCl, 2.5 mM MgCl2, and double-distilled water [ddH2O]) for 45 min at 25°C. Subsequently, samples were rinsed and washed five times for 3 min with 50 mM cacodylate buffer. Next, samples were fixed for 90–120 min with 2% OsO4 in 50 mM cacodylate buffer and shifted to aqueous media by washing five times in short intervals in distilled water. Then, samples were contrasted overnight in aqueous 0.5% uranyl acetate. After five washes with ddH2O, the samples were dehydrated in an ethanol series of 50%, 70%, 90%, and 96% and three times at 100% for 15 min each. In preparation of tissue embedding, samples were washed two times for 20 min with propylene oxide. Thereafter, the tissue was carefully infiltrated and embedded in conventional epon, closed with a gelatin cap, and cured at 60°C for 48 h. Subsequently, 60–80-nm ultrathin sections were contrasted for 8 min using 5% filtered uranyl acetate in ethanol. Between and after incubation steps, sections were dipped into 100% ethanol, 50% ethanol, and ddH2O. After careful drying, the sections were again contrasted with Reynolds' lead citrate (for 4 min; Reynolds, 1963) in decocted ddH2O and washed three times in decocted ddH2O. Image acquisition was performed with a Zeiss EM 900. Images were registered on photo plates. The negatives were scanned (1,200 dpi) to digitalize the images for subsequent analysis. The number of SVs at a particular synapse was quantified within four 50-nm shells (in nm: 50–100, 100–150, 150–200, and 200–250) surrounding the T-bar (Hallermann et al., 2010b). Micrographs were acquired with 85,000-fold magnification.
Mouse RB terminal
The preparation of retinae from 8–10-wk-old cpx3KO and WT mice has been described previously (Mortensen et al., 2016, Supplemental Experimental Procedures). Briefly, retinae were fixed in a cocktail of 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer, embedded in low-melt agarose, and sectioned into 100-µm slices using a vibratome. Retina slices were processed by high-pressure freezing (Leica HPM100) and automated freeze substitution (Leica EM AFS2) and embedded in plastic for ultramicrotomy. Electron micrographs were acquired on a transmission electron microscope (Zeiss LEO 912-Omega, 80 kV) with 25,000-fold magnification. SVs surrounding ribbons of WT and cpx3KO RB synapses were counted within 200-nm concentric shells (in nm: 0–200, 200–400, 400–600, 600–800, and 800–1,000) centered at the ribbon base. The cytoplasmic area covered by each shell was also quantified to take into account cell morphology at presynaptic sites. In Fig. 6, image contrast was manually adjusted to ease visualization.
Synthesis of Brp peptides and interaction assays
For affinity purification of Cpx, a 31–amino acid peptide (CTSVVPFPGGGGGQQQQQQDAGPAGFLKSFF) including BrpC-tip (underlined) and a scrambled control peptide (CTSVVPFPGGGGGPFQQSLGFKAQQAQDQFG) was synthesized by standard solid-phase peptide synthesis using Fmoc chemistry. Peptides were immobilized to iodoacetyl agarose via the artificial cysteine residue added to their N terminus. For photoaffinity labeling of Cpx, a photoreactive variant of the BrpC-tip peptide, was synthesized by replacement of the Phe residue in position 26 with para-benzoyl-Phe (Bpa) as described previously (Jahn et al., 2002). For detection purposes, the photopeptide was additionally equipped with a biotin moiety via maleimide coupling (Maleimide-PEG2-Biotin; Thermo Fisher Scientific) to the sulfhydryl group of the N-terminal Cys residue, the product referred to as Biotin-Brp-Bpa peptide.
Drosophila heads (from ∼30,000 w1118 animals) were homogenized in solubilization buffer (150 mM NaCl, 10 mM Hepes-NaOH, pH 7.4, 1 mM EGTA, 2 mM MgCl2, 1% NP-40, 1 mM DTT, and protease inhibitor cocktail) using an Ultra-Turrax (IKA), solubilized for 20 min (4°C), and centrifuged at 16,000 g for 30 min at 4°C. Next, the supernatant was collected into a fresh tube and proteins solubilized for another 10 min at 4°C before 15 min ultracentrifugation at 346,000 g. For the affinity purification approach, the resulting supernatant 2 was mixed with 100-µl functionalized beads and incubated for 3 h at 4°C. Subsequently, beads were washed five times using solubilization buffer, resuspended in 1× Laemmli sample buffer and boiled for 5 min. For the photoaffinity labeling approach, supernatant 2 was mixed with Biotin-Brp-Bpa peptide (10 µM final concentration) and incubated for 3 h at 4°C in the absence or presence of an excess of unmodified BrpC-tip peptide (100 µM final concentration; referred to as WT Brp peptide). Subsequently, samples were exposed to UV light for 20 min on ice as described elsewhere (Jahn et al., 2002) and subjected to gel electrophoresis followed by immunoblotting.
Western blots
Fly heads were collected in standard radioimmunoprecipitation assay buffer (RIPA buffer; 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) supplemented with protease inhibitor cocktail (1:1,000; Sigma-Aldrich) and immediately frozen in liquid nitrogen. Next, heads were homogenized using a pipette tip, supplemented with SDS-based protein buffer (Li-cor) and 2-mercaptoethanol (Merck). After a brief vortexing step, samples were centrifuged for 5 min at 13,000 rpm (4°C), incubated for 10 min at 55°C, subjected to electrophoresis on a 4–12% SDS gel, and blotted onto a 0.2-µm nitrocellulose membrane (Amersham Protran). The membrane was blocked for 1 h using Odyssey Blocking buffer (Li-cor) diluted 1:8 with 1xPBS. Blots were probed with primary antisera at the indicated concentration for 1 h at RT (rabbit-α-Cpx; 1:10,000; Huntwork and Littleton, 2007; and mouse-α-tubulinβ; 1:1,000; e7, RRID: AB_528499; Developmental Studies Hybridoma Bank). After rinsing twice and undergoing three 10-min washing steps, membranes were incubated with IRDye 680RD goat-α-rabbit (RRID: AB_10956166) and 800CW goat-α-mouse (1:16,000, RRID: AB_621842; Li-cor) for 1 h at RT, rinsed twice, and washed three times for 10 min. Proteins extracted from w1118 fly head homogenate via affinity purification experiments were separated by SDS-PAGE (18% gel), immunoblotted using rabbit-α-Cpx (1:10,000, RRID: AB_2568068; Huntwork and Littleton, 2007), and goat-α-rabbit Alexa Fluor 680 secondary antibody (1:5,000; Life). Protein fractions derived from photoaffinity labeling experiments were separated as above or on precast Bis-Tris NuPAGE 4–12% gradient gels (Thermo Fisher Scientific) with MES running buffer. Visualization of Cpx was as above, while biotin was coimmunodetected by Streptavidin IRDye 800CW (1:2,500; Rockland). Western blots were documented with a near-infrared imager (Odyssey; Li-cor).
Electrophysiology
TEVC recordings (Axoclamp 900A amplifier; Molecular Devices) were made from muscle 6, segments A2 and A3 of late third-instar male Drosophila larvae essentially as previously described (Ljaschenko et al., 2013). All measurements were obtained at RT in HL-3 with the following composition (in mM): NaCl 70, KCl 5, MgCl2 20, NHCO3 10, trehalose 5, sucrose 115, Hepes 5, and CaCl2 1.5, 1, or 0.6 (as indicated), pH adjusted to 7.2. The intracellular electrodes had resistances of 10–20 MΩ, filled with 3 M KCl. For analysis, only cells with an initial membrane potential of at least −50 mV and a membrane resistance of ≥4 MΩ were included. During recordings, cells were clamped at a holding potential of −80 mV (minis) or −60 mV (eEPSCs). To evoke synaptic currents, nerves were stimulated via a suction electrode (diameter ∼15 µm) with 300-µs pulses, typically at 10 V (Grass S88 stimulator and isolation unit SIU5; Astro-Med). Signals were low-pass filtered at 10 kHz and analyzed in Clampfit 10.2 (Molecular Devices). Paired-pulse recordings were done with interstimulus intervals of (in ms: 10, 30, 100, 300, and 1,000). Between recordings, cells were given a 10-s rest. For analysis, 10 traces per interval were averaged. The amplitude of the second response in 10-ms interpulse recordings was measured from the peak to the point of interception with the extrapolated first eEPSC. To measure τ decay, the decaying phase of evoked currents was fitted with a monoexponential function from 60% of the peak amplitude to the end of the event. To estimate RRV pool sizes, a train of 100 pulses was applied at 60 Hz. Linear fits from 0.3 to 0.5 s were applied to cumulatively plotted eEPSCs and back extrapolated (Hallermann et al., 2010a; Weyhersmüller et al., 2011).
Modeling
We used a model with two pools of RRVs and heterogeneous release probabilities similar to that described in Ehmann et al. (2014). The model consisted of two pools of RRVs (N1 and N2) with release probabilities pvr1 and pvr2, respectively, and a supply pool N0. N2 is refilled with rate k2 from N1, N1 is refilled with rate k1 from N0, and N0 is refilled with rate k0 from an infinite reserve pool of SVs. Synaptic facilitation was implemented according to Markram et al. (1998). To prevent overdetermination (i.e., to obtain results independent of the start values), the model had five free parameters (N0, N2, k1, pvr1, and pvr2), and the remaining parameters were constrained (N1 = 4*N2, k2 = 0.15 s−1, k0 = 0.03 s−1).
Larval locomotion
Briefly, wandering third-instar larvae were positioned in a Petri dish (9 cm in diameter) filled with 1% agarose. The crawling paths of each genotype were recorded for 2 min using a digital camera, and path lengths of individual animals were subsequently measured in ImageJ.
Statistics
Data were analyzed with Prism 5.0 (GraphPad) and Sigma Plot 12.5 (Software Inc.). Group means were compared by a two-tailed t test, unless the assumption of normality of the sample distribution was violated. In this case, group means were compared by a nonparametric rank sum test. Data are reported as mean ± SEM; n indicates the sample number, and P denotes the level of significance (*, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001). Statistics of experiments are summarized in Tables S1, S2, and S4–S11.
Online supplemental material
Fig. S1 provides information on SV tethering by BrpC-tip and Cpx expression levels upon RNAi. Fig. S2 shows the subcellular distribution of Cpx in the cpx1257 mutant. Fig. S3 displays the results of biochemical interaction studies. Fig. S4 shows electrophysiological data of single mutants and DMs and SIM images of cpx1257 NMJs. Fig. S5 shows that cpx3KO ribbons are surrounded by a normal cytoplasmic area in mouse RB terminals. Table S1 summarizes data on larval locomotion. Table S2 shows paired-pulse ratio values upon BrpC-tip expression. Table S3 lists the candidate genes, and Table S4 provides imaging values for the genetic screen. Table S5 summarizes the EM data on SV tethering to T-bars, and Table S6 displays dSTORM values for CAZ measurements. Table S7 shows the electrophysiological data. Table S8 provides values for the confocal quantification of active zone numbers, and Table S9 shows numerical results for RRV pool estimates. Tables S10 and S11 show imaging results for Ca2+ channel clusters via SIM and synaptic ribbons via EM, respectively.
Supplementary Material
Acknowledgments
We thank T. Littleton for sharing the Cpx antibody, F. Kawasaki for cpx1257 flies, L. van Werven, T. Hellmann, M. Oppmann, A. Dahse and B. Trost for expert technical assistance, C. Wichmann for ultrastructural advice, E. Buchner for discussions, and M. Sauer for supporting SIM experiments.
This work was funded by the Deutsche Forschungsgemeinschaft (grant FOR 2149/P01 to N. Scholz; grants FOR 2149/P01 and FOR 2149/P03, SFB 1047/A05, TRR 166/C03, and LA2861/7-1 to T. Langenhan; and grants FOR 2149/P03, SFB 1047/A05, TRR 166/B04, and KI1460/4-1 to R.J. Kittel). N. Ehmann was supported by the German Excellence Initiative (grant GSC106/3 to the Graduate School of Life Sciences, University of Würzburg [PostDoc Plus program]). N. Scholz was supported by the Medical Faculty, University Leipzig (Junior Research grant).
The authors declare no competing financial interests.
Author contributions: N. Scholz, N. Ehmann, T. Langenhan, and R.J. Kittel conceived the study; N. Scholz, N. Ehmann, D. Sachidanandan, C. Imig, B.H. Cooper, O. Jahn, K. Reim, J. Meyer, M. Lamberty, S. Altrichter, A. Bormann, S. Hallermann, C. Stigloher, T. Langenhan, and R.J. Kittel performed investigations, data curation, and analysis; N. Scholz, N. Ehmann, S. Hallermann, T. Langenhan, and R.J. Kittel created models; N. Scholz, N. Ehmann, N. Brose, S. Hallermann, M. Pauli, M. Heckmann, C. Stigloher, T. Langenhan, and R.J. Kittel provided resources; S. Hallermann carried out programming; N. Brose, C. Stigloher, T. Langenhan, and R.J. Kittel provided supervision; N. Scholz, N. Ehmann, O. Jahn, K. Reim, S. Hallermann, T. Langenhan, and R.J. Kittel validated the experimental results and visualized the data together with D. Sachidanandan and M. Lamberty; T. Langenhan, and R.J. Kittel administered the project and wrote the manuscript; and all authors participated in manuscript review and editing.
References
- Babai N., Sendelbeck A., Regus-Leidig H., Fuchs M., Mertins J., Reim K., Brose N., Feigenspan A., and Brandstätter J.H.. 2016. Functional roles of Complexin3 and Complexin4 at mouse photoreceptor ribbon synapses. J. Neurosci. 36:6651–6667. 10.1523/JNEUROSCI.4335-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H., and Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Brose N. 2008. For better or for worse: complexins regulate SNARE function and vesicle fusion. Traffic. 9:1403–1413. 10.1111/j.1600-0854.2008.00758.x [DOI] [PubMed] [Google Scholar]
- Buhl L.K., Jorquera R.A., Akbergenova Y., Huntwork-Rodriguez S., Volfson D., and Littleton J.T.. 2013. Differential regulation of evoked and spontaneous neurotransmitter release by C-terminal modifications of complexin. Mol. Cell. Neurosci. 52:161–172. 10.1016/j.mcn.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R.W., Song Y., and Littleton J.T.. 2010. Comparative analysis of Drosophila and mammalian complexins as fusion clamps and facilitators of neurotransmitter release. Mol. Cell. Neurosci. 45:389–397. 10.1016/j.mcn.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B.J., Imlach W.L., Jiao W., Wolfram V., Wu Y., Grbic M., Cela C., Baines R.A., Nitabach M.N., and McCabe B.D.. 2014. Miniature neurotransmission regulates Drosophila synaptic structural maturation. Neuron. 82:618–634. 10.1016/j.neuron.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford D.C., and Kavalali E.T.. 2015. Molecular underpinnings of synaptic vesicle pool heterogeneity. Traffic. 16:338–364. 10.1111/tra.12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R.W., Collins C.A., Gelfand M.V., Dant J., Brooks E.S., Krantz D.E., and DiAntonio A.. 2004. Increased expression of the Drosophila vesicular glutamate transporter leads to excess glutamate release and a compensatory decrease in quantal content. J. Neurosci. 24:10466–10474. 10.1523/JNEUROSCI.3001-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R.W., Gelfand M.V., Collins C.A., and DiAntonio A.. 2008. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J. Comp. Neurol. 508:131–152. 10.1002/cne.21670 [DOI] [PubMed] [Google Scholar]
- Denker A., Krohnert K., Buckers J., Neher E., and Rizzoli S.O.. 2011. The reserve pool of synaptic vesicles acts as a buffer for proteins involved in synaptic vesicle recycling. Proc. Natl. Acad. Sci. USA. 108:17183–17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A., Petersen S.A., Heckmann M., and Goodman C.S.. 1999. Glutamate receptor expression regulates quantal size and quantal content at the Drosophila neuromuscular junction. J. Neurosci. 19:3023–3032. 10.1523/JNEUROSCI.19-08-03023.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. . 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Ehmann N., van de Linde S., Alon A., Ljaschenko D., Keung X.Z., Holm T., Rings A., DiAntonio A., Hallermann S., Ashery U., et al. . 2014. Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 5:4650 10.1038/ncomms5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Busnadiego R., Asano S., Oprisoreanu A.M., Sakata E., Doengi M., Kochovski Z., Zürner M., Stein V., Schoch S., Baumeister W., and Lucić V.. 2013. Cryo-electron tomography reveals a critical role of RIM1α in synaptic vesicle tethering. J. Cell Biol. 201:725–740. 10.1083/jcb.201206063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet W., Owald D., Wichmann C., Mertel S., Depner H., Dyba M., Hallermann S., Kittel R.J., Eimer S., and Sigrist S.J.. 2009. Maturation of active zone assembly by Drosophila Bruchpilot. J. Cell Biol. 186:129–145. 10.1083/jcb.200812150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank T., Rutherford M.A., Strenzke N., Neef A., Pangršič T., Khimich D., Fejtova A., Gundelfinger E.D., Liberman M.C., Harke B., et al. . 2010. Bassoon and the synaptic ribbon organize Ca2+ channels and vesicles to add release sites and promote refilling. Neuron. 68:724–738. 10.1016/j.neuron.2010.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Lai Y., Li X., Wang M., Leitz J., Hu Y., Zhang Y., Choi U.B., Cipriano D., Pfuetzner R.A., et al. . 2016. C-terminal domain of mammalian complexin-1 localizes to highly curved membranes. Proc. Natl. Acad. Sci. USA. 113:E7590–E7599. 10.1073/pnas.1609917113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A.C., Fish M., Nusse R., and Calos M.P.. 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 166:1775–1782. 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M.G.L., Shao L., Carlton P.M., Wang C.J.R., Golubovskaya I.N., Cande W.Z., Agard D.A., and Sedat J.W.. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallermann S., and Silver R.A.. 2013. Sustaining rapid vesicular release at active zones: potential roles for vesicle tethering. Trends Neurosci. 36:185–194. 10.1016/j.tins.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallermann S., Fejtova A., Schmidt H., Weyhersmüller A., Silver R.A., Gundelfinger E.D., and Eilers J.. 2010c Bassoon speeds vesicle reloading at a central excitatory synapse. Neuron. 68:710–723. 10.1016/j.neuron.2010.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallermann S., Heckmann M., and Kittel R.J.. 2010a Mechanisms of short-term plasticity at neuromuscular active zones of Drosophila. HFSP J. 4:72–84. 10.2976/1.3338710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallermann S., Kittel R.J., Wichmann C., Weyhersmüller A., Fouquet W., Mertel S., Owald D., Eimer S., Depner H., Schwärzel M., et al. . 2010b Naked dense bodies provoke depression. J. Neurosci. 30:14340–14345. 10.1523/JNEUROSCI.2495-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann M., and Dudel J.. 1997. Desensitization and resensitization kinetics of glutamate receptor channels from Drosophila larval muscle. Biophys. J. 72:2160–2169. 10.1016/S0006-3495(97)78859-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M., van de Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., and Sauer M.. 2008. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47:6172–6176. 10.1002/anie.200802376 [DOI] [PubMed] [Google Scholar]
- Hobson R.J., Liu Q., Watanabe S., and Jorgensen E.M.. 2011. Complexin maintains vesicles in the primed state in C. elegans. Curr. Biol. 21:106–113. 10.1016/j.cub.2010.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi N., Holt M., and Sakaba T.. 2009. Calcium dependence of exo- and endocytotic coupling at a glutamatergic synapse. Neuron. 63:216–229. 10.1016/j.neuron.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Huntwork S., and Littleton J.T.. 2007. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat. Neurosci. 10:1235–1237. 10.1038/nn1980 [DOI] [PubMed] [Google Scholar]
- Imig C., Min S.-W., Krinner S., Arancillo M., Rosenmund C., Südhof T.C., Rhee J., Brose N., and Cooper B.H.. 2014. The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron. 84:416–431. 10.1016/j.neuron.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., and Yamamoto D.. 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 124:761–771. [DOI] [PubMed] [Google Scholar]
- Iyer J., Wahlmark C.J., Kuser-Ahnert G.A., and Kawasaki F.. 2013. Molecular mechanisms of COMPLEXIN fusion clamp function in synaptic exocytosis revealed in a new Drosophila mutant. Mol. Cell. Neurosci. 56:244–254. 10.1016/j.mcn.2013.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O., Eckart K., Brauns O., Tezval H., and Spiess J.. 2002. The binding protein of corticotropin-releasing factor: ligand-binding site and subunit structure. Proc. Natl. Acad. Sci. USA. 99:12055–12060. 10.1073/pnas.192449299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch W.J., Speidel D., Sigler A., Sørensen J.B., Varoqueaux F., Rhee J.S., and Brose N.. 2007. CAPS-1 and CAPS-2 are essential synaptic vesicle priming proteins. Cell. 131:796–808. 10.1016/j.cell.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Kaeser P.S., and Regehr W.G.. 2014. Molecular mechanisms for synchronous, asynchronous, and spontaneous neurotransmitter release. Annu. Rev. Physiol. 76:333–363. 10.1146/annurev-physiol-021113-170338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F., Hazen M., and Ordway R.W.. 2000. Fast synaptic fatigue in shibire mutants reveals a rapid requirement for dynamin in synaptic vesicle membrane trafficking. Nat. Neurosci. 3:859–860. 10.1038/78753 [DOI] [PubMed] [Google Scholar]
- Kawasaki F., Zou B., Xu X., and Ordway R.W.. 2004. Active zone localization of presynaptic calcium channels encoded by the cacophony locus of Drosophila. J. Neurosci. 24:282–285. 10.1523/JNEUROSCI.3553-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel R.J., Wichmann C., Rasse T.M., Fouquet W., Schmidt M., Schmid A., Wagh D.A., Pawlu C., Kellner R.R., Willig K.I., et al. . 2006. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 312:1051–1054. 10.1126/science.1126308 [DOI] [PubMed] [Google Scholar]
- Ljaschenko D., Ehmann N., and Kittel R.J.. 2013. Hebbian plasticity guides maturation of glutamate receptor fields in vivo. Cell Reports. 3:1407–1413. 10.1016/j.celrep.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Markram H., Wang Y., and Tsodyks M.. 1998. Differential signaling via the same axon of neocortical pyramidal neurons. Proc. Natl. Acad. Sci. USA. 95:5323–5328. 10.1073/pnas.95.9.5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S.E., and Perrimon N.. 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40:476–483. 10.1038/ng.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.A., Hu Z., Fenz K.M., Fernandez J., and Dittman J.S.. 2011. Complexin has opposite effects on two modes of synaptic vesicle fusion. Curr. Biol. 21:97–105. 10.1016/j.cub.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melom J.E., Akbergenova Y., Gavornik J.P., and Littleton J.T.. 2013. Spontaneous and evoked release are independently regulated at individual active zones. J. Neurosci. 33:17253–17263. 10.1523/JNEUROSCI.3334-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa M., and Sakaba T.. 2015. Imaging exocytosis of single synaptic vesicles at a fast CNS presynaptic terminal. Neuron. 88:492–498. 10.1016/j.neuron.2015.09.047 [DOI] [PubMed] [Google Scholar]
- Miskiewicz K., Jose L.E., Yeshaw W.M., Valadas J.S., Swerts J., Munck S., Feiguin F., Dermaut B., and Verstreken P.. 2014. HDAC6 is a Bruchpilot deacetylase that facilitates neurotransmitter release. Cell Reports. 8:94–102. 10.1016/j.celrep.2014.05.051 [DOI] [PubMed] [Google Scholar]
- Mortensen L.S., Park S.J.H., Ke J.B., Cooper B.H., Zhang L., Imig C., Löwel S., Reim K., Brose N., Demb J.B., et al. . 2016. Complexin 3 increases the fidelity of signaling in a retinal circuit by regulating exocytosis at ribbon synapses. Cell Reports. 15:2239–2250. 10.1016/j.celrep.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. 2010a Complexin: does it deserve its name? Neuron. 68:803–806. 10.1016/j.neuron.2010.11.038 [DOI] [PubMed] [Google Scholar]
- Neher E. 2010b What is rate-limiting during sustained synaptic activity: vesicle supply or the availability of release sites. Front. Synaptic Neurosci. 2:144 10.3389/fnsyn.2010.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. 2015. Merits and limitations of vesicle pool models in view of heterogeneous populations of synaptic vesicles. Neuron. 87:1131–1142. 10.1016/j.neuron.2015.08.038 [DOI] [PubMed] [Google Scholar]
- Paul M.M., Pauli M., Ehmann N., Hallermann S., Sauer M., Kittel R.J., and Heckmann M.. 2015. Bruchpilot and Synaptotagmin collaborate to drive rapid glutamate release and active zone differentiation. Front. Cell. Neurosci. 9:29 10.3389/fncel.2015.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim K. 2017. Complexins. In Reference Module in Neuroscience and Biobehavioral Psychology. Elsevier Inc. 10.1016/B978-0-12-809324-5.02507-4 [DOI] [Google Scholar]
- Reim K., Wegmeyer H., Brandstätter J.H., Xue M., Rosenmund C., Dresbach T., Hofmann K., and Brose N.. 2005. Structurally and functionally unique complexins at retinal ribbon synapses. J. Cell Biol. 169:669–680. 10.1083/jcb.200502115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim K., Regus-Leidig H., Ammermüller J., El-Kordi A., Radyushkin K., Ehrenreich H., Brandstätter J.H., and Brose N.. 2009. Aberrant function and structure of retinal ribbon synapses in the absence of complexin 3 and complexin 4. J. Cell Sci. 122:1352–1361. 10.1242/jcs.045401 [DOI] [PubMed] [Google Scholar]
- Reynolds E.S. 1963. The use of lead citrate stain at high pH in electron microscopy. J. Cell Biol. 17:208–212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoli S.O., and Betz W.J.. 2004. The structural organization of the readily releasable pool of synaptic vesicles. Science. 303:2037–2039. 10.1126/science.1094682 [DOI] [PubMed] [Google Scholar]
- Robinson S.W., Bourgognon J.-M., Spiers J.G., Breda C., Campesan S., Butcher A., Mallucci G.R., Dinsdale D., Morone N., Mistry R., et al. . 2018. Nitric oxide-mediated posttranslational modifications control neurotransmitter release by modulating complexin farnesylation and enhancing its clamping ability. PLoS Biol. 16:e2003611 10.1371/journal.pbio.2003611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeva N., Cho R.W., Vasin A., Gonzalez A., Littleton J.T., and Bykhovskaia M.. 2017. Complexin mutants reveal partial segregation between recycling pathways that drive evoked and spontaneous neurotransmission. J. Neurosci. 37:383–396. 10.1523/JNEUROSCI.1854-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S. 2009. Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr. Patterns. 9:371–380. 10.1016/j.gep.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Schäfer P., van de Linde S., Lehmann J., Sauer M., and Doose S.. 2013. Methylene blue- and thiol-based oxygen depletion for super-resolution imaging. Anal. Chem. 85:3393–3400. 10.1021/ac400035k [DOI] [PubMed] [Google Scholar]
- Schmid A., Hallermann S., Kittel R.J., Khorramshahi O., Frölich A.M.J., Quentin C., Rasse T.M., Mertel S., Heckmann M., and Sigrist S.J.. 2008. Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat. Neurosci. 11:659–666. 10.1038/nn.2122 [DOI] [PubMed] [Google Scholar]
- Schneggenburger R., Meyer A.C., and Neher E.. 1999. Released fraction and total size of a pool of immediately available transmitter quanta at a calyx synapse. Neuron. 23:399–409. 10.1016/S0896-6273(00)80789-8 [DOI] [PubMed] [Google Scholar]
- Sharonov A., and Hochstrasser R.M.. 2007. Single-molecule imaging of the association of the cell-penetrating peptide Pep-1 to model membranes. Biochemistry. 46:7963–7972. 10.1021/bi700505h [DOI] [PubMed] [Google Scholar]
- Snellman J., Mehta B., Babai N., Bartoletti T.M., Akmentin W., Francis A., Matthews G., Thoreson W., and Zenisek D.. 2011. Acute destruction of the synaptic ribbon reveals a role for the ribbon in vesicle priming. Nat. Neurosci. 14:1135–1141. 10.1038/nn.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Stewart B.A., Atwood H.L., Renger J.J., Wang J., and Wu C.F.. 1994. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 175:179–191. 10.1007/BF00215114 [DOI] [PubMed] [Google Scholar]
- Südhof T.C. 2012. The presynaptic active zone. Neuron. 75:11–25. 10.1016/j.neuron.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., and Sakata-Sogawa K.. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 5:159–161. 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- Trimbuch T., and Rosenmund C.. 2016. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat. Rev. Neurosci. 17:118–125. 10.1038/nrn.2015.16 [DOI] [PubMed] [Google Scholar]
- van de Linde S., Löschberger A., Klein T., Heidbreder M., Wolter S., Heilemann M., and Sauer M.. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6:991–1009. 10.1038/nprot.2011.336 [DOI] [PubMed] [Google Scholar]
- Wagh D.A., Rasse T.M., Asan E., Hofbauer A., Schwenkert I., Dürrbeck H., Buchner S., Dabauvalle M.C., Schmidt M., Qin G., et al. . 2006. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 49:833–844. 10.1016/j.neuron.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Wang S., Yoo S., Kim H.-Y., Wang M., Zheng C., Parkhouse W., Krieger C., and Harden N.. 2015. Detection of in situ protein-protein complexes at the Drosophila larval neuromuscular junction using proximity ligation assay. J. Vis. Exp. 95:52139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.S.H., Held R.G., Wong M.Y., Liu C., Karakhanyan A., and Kaeser P.S.. 2016. Fusion competent synaptic vesicles persist upon active zone disruption and loss of vesicle docking. Neuron. 91:777–791. 10.1016/j.neuron.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyhersmüller A., Hallermann S., Wagner N., and Eilers J.. 2011. Rapid active zone remodeling during synaptic plasticity. J. Neurosci. 31:6041–6052. 10.1523/JNEUROSCI.6698-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B.G., Mandad S., Truckenbrodt S., Kröhnert K., Schäfer C., Rammner B., Koo S.J., Claßen G.A., Krauss M., Haucke V., et al. . 2014. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 344:1023–1028. 10.1126/science.1252884 [DOI] [PubMed] [Google Scholar]
- Wolter S., Schüttpelz M., Tscherepanow M., van de Linde S., Heilemann M., and Sauer M.. 2010. Real-time computation of subdiffraction-resolution fluorescence images. J. Microsc. 237:12–22. 10.1111/j.1365-2818.2009.03287.x [DOI] [PubMed] [Google Scholar]
- Wolter S., Löschberger A., Holm T., Aufmkolk S., Dabauvalle M.C., van de Linde S., and Sauer M.. 2012. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods. 9:1040–1041. 10.1038/nmeth.2224 [DOI] [PubMed] [Google Scholar]
- Wragg R.T., Snead D., Dong Y., Ramlall T.F., Menon I., Bai J., Eliezer D., and Dittman J.S.. 2013. Synaptic vesicles position complexin to block spontaneous fusion. Neuron. 77:323–334. 10.1016/j.neuron.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Lin Y.Q., Pan H., Reim K., Deng H., Bellen H.J., and Rosenmund C.. 2009. Tilting the balance between facilitatory and inhibitory functions of mammalian and Drosophila Complexins orchestrates synaptic vesicle exocytosis. Neuron. 64:367–380. 10.1016/j.neuron.2009.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K.J., Proulx S.R., and Jorgensen E.M.. 2001. Rules of nonallelic noncomplementation at the synapse in Caenorhabditis elegans. Genetics. 158:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenisek D., Steyer J.A., and Almers W.. 2000. Transport, capture and exocytosis of single synaptic vesicles at active zones. Nature. 406:849–854. 10.1038/35022500 [DOI] [PubMed] [Google Scholar]
- Zhai R.G., and Bellen H.J.. 2004. The architecture of the active zone in the presynaptic nerve terminal. Physiology. 19:262–270. 10.1152/physiol.00014.2004 [DOI] [PubMed] [Google Scholar]
- Zhang F.L., and Casey P.J.. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65:241–269. 10.1146/annurev.bi.65.070196.001325 [DOI] [PubMed] [Google Scholar]
- Zucker R.S., and Regehr W.G.. 2002. Short-term synaptic plasticity. Annu. Rev. Physiol. 64:355–405. 10.1146/annurev.physiol.64.092501.114547 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.