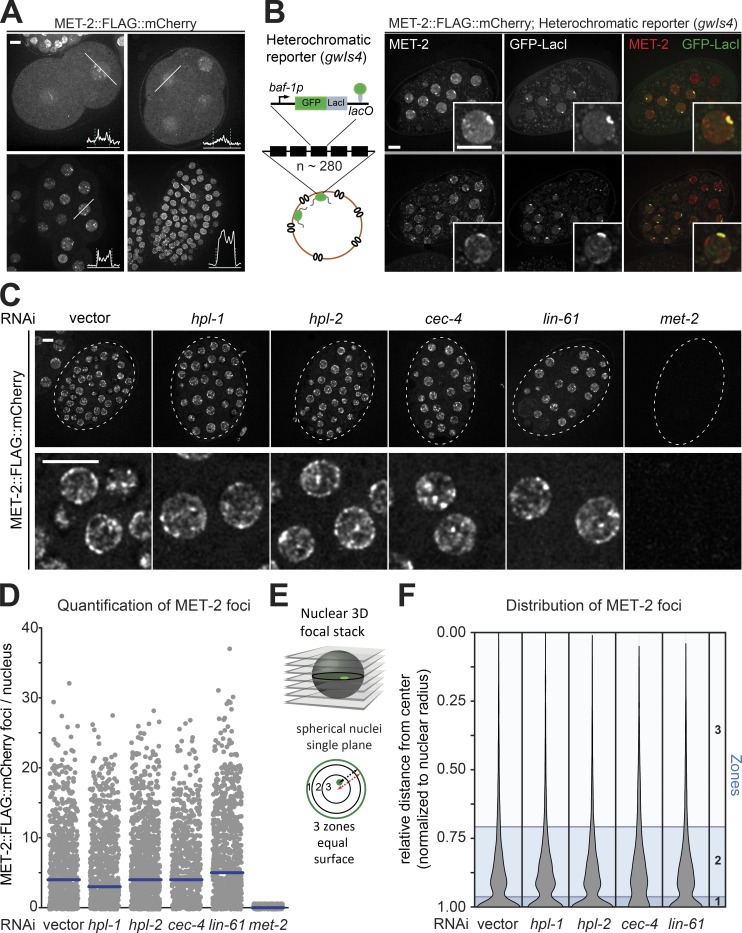
Figure 2.
Integrity and prevalence of MET-2 foci are independent of canonical H3K9me readers. (A) Live-cell images of 2-, 4-, 20-, and ∼100-cell embryos expressing MET-2::FLAG::mCherry, with line profiles averaged over a 10-pixel-width zone for mCherry intensity. Dashed lines indicate peaks of LEM-2::GFP signal. Bar = 5 µm. (B) Live-cell images of embryos expressing met-2::flag::mcherry and gfp::lacI from the integrated heterochromatic array (∼280 copies) of gwIs4 [baf-1p::GFP-lacI::let-858 3′UTR; myo-3p::RFP]. Assay visualized through GFP-lacI binding to lacO site (Meister et al., 2010b). (C) Example images of MET-2::FLAG::mCherry in 50-cell-stage embryos treated with RNAi against hpl-1, hpl-2, cec-4, lin-61, met-2, or empty vector (vector). The boxed region is enlarged in the lower row. Bar = 5 µm. (D) Quantitation of MET-2::FLAG::mCherry foci in the embryos shown in C using automated image analysis. N = 3, nuclei scored: n (vector) = 873, n (hpl-1) = 948, n (hpl-2) = 1,043, n (cec-4) = 675, n (lin-61) = 881, and n (met-2) = 589. (E) Scheme of nuclear zoning assay (Meister et al., 2010a). See Materials and methods for explanation of zones; zone 1 is the most peripheral. (F) Violin plot showing the cumulative spread of distances of MET-2::FLAG::mCherry foci to the nuclear periphery. Each measurement is normalized to the radius. Zones calculated as in E are shown in blue. N = 3, nuclei scored: n (vector) = 4,822, n (hpl-1) = 4,852, n (hpl-2) = 6,108, n (cec-4) = 3,991, and n (lin-61) = 6,126.