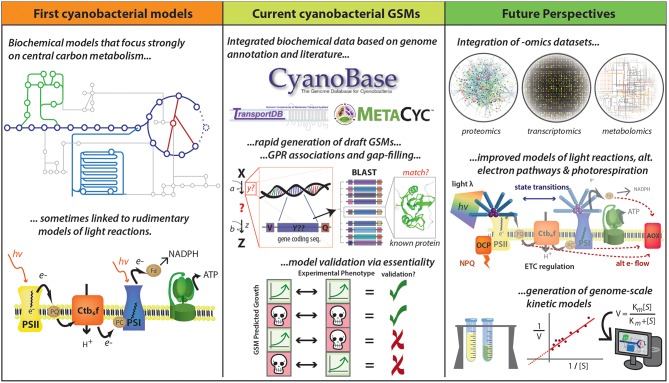
Figure 4.
Development of cyanobacterial GSMs. Early cyanobacterial GSMs were developed based on biochemical data and were focused mainly on central carbon metabolism and linked to basic representations of photosynthetic pathways (Cogne et al., 2003; Shastri and Morgan, 2005; Hong and Lee, 2007). Numerous advances have accelerated the reconstruction of GSMs, including improved quantity and quality of data described in public databases (e.g., CyanoBase, TransportDB, KEGG, and MetaCyc) and the development of tools for rapid generation of drafts and for gap-filling. The quality of GSMs has been substantially improved with the inclusion of gene-protein-reaction (GPR) associations, which has been assessed by essential gene prediction based on the availability of experimental data. This methodology allows identify four different kind of predictions: true positives, true negatives, false positives, and false negatives. True positives and true negatives are obtained when both prediction and experimental data indicate a gene is essential and non-essential, respectively (the first and the second case). False positives occur when the model says a gene is essential, but experiments suggest otherwise (the third case). False negatives are generated when a gene is predicted to be non-essential but in reality, it is essential (the fourth case). However, new improvements could further improve GSM predictive ability, including the integration of—omics datasets and the improvement of photosynthesis and photorespiration modeling. A long-term goal would be the development of genome-scale kinetic models, which might be expected to provide more accurate metabolic predictions. Logos for MetaCyc, TransportDB, and CyanoBase are used with permission.