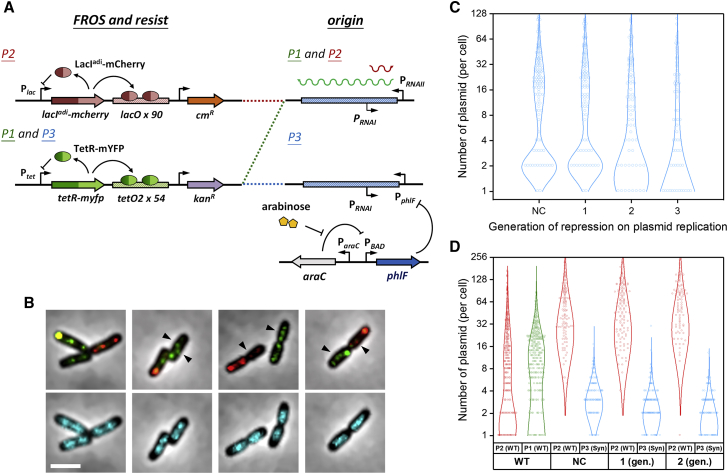
Figure 1.
Single hcn plasmid molecules can be resolved by two-color FROS and copy-number control systems. (A) The design schemes of the self-regulated TetR-YFP/tetO and LacIadi-mCherry/lacO systems are shown. Antibiotic resistance and replication origins, both wt (pTetORK34b (P1) and pLacOIC2c (P2)) and synthetic (pTetORK34p (P3)), are also shown. LacIadi is a LacI mutant that can only form dimers, not tetramers. (B) Photomicrographs show phase contrast images of cells merged with fluorescence images of cotransformed plasmids (P1 (upper panel, green) and P2 (upper panel, red)) or nucleoids (lower panel, cyan). The arrows indicate probable single plasmids that are traveling around the nucleoids. Scale bars, 3 μm. Violin plots show the estimated numbers of plasmids in cells according to generations of repression. (C) Cells were transformed with only P3 (cyan), or (D) cells were additionally transformed with the wt plasmid, P2 (red). The estimated plasmid numbers without repression are presented as the negative control (NC), and those in cells cotransformed with P1 (green) and P2 are also shown for comparison. The circles and lines are histograms and Gaussian fits, respectively. The numbers of cells in the analyzed in (C) are 151, 127, 88, and 87 from left to right. In (D), the numbers of cells are 290, 124, 136, and 103 from left to right. Of note, the asymmetry between P1 and P2 may be due to differences in antibiotic selection. To see this figure in color, go online.