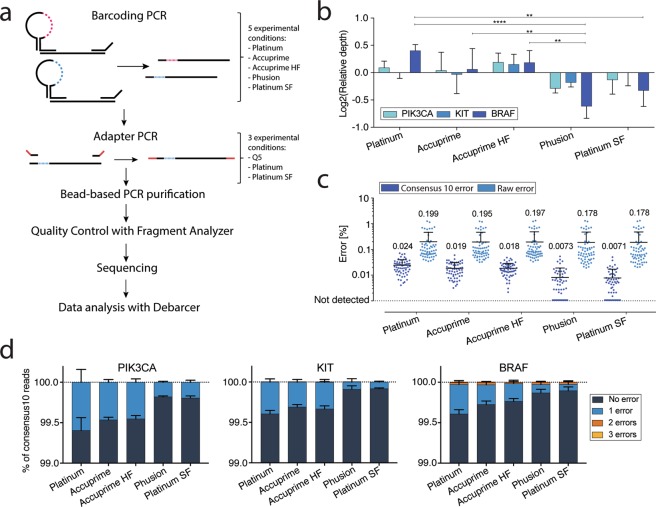
Figure 1.
Effect of polymerase fidelity in the initial barcoding PCR. (a) Workflow for SimSen-Seq indicating which polymerases were tested in the barcoding and adapter PCR steps. Dotted lines represent barcodes with colors indicating different barcode sequences. Red lines indicate the P5 and P7 adapter sequences. (b) Relative consensus10 sequencing depths for each amplicon using five different polymerases in the barcoding PCR step (Q5 polymerase was used for all adapter PCRs). Consensus depths were compared using 2-way ANOVA (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). (c) Comparison of raw and consensus10 errors observed at all base positions (all three amplicons combined) using five different polymerases in the barcoding PCR and Q5 polymerase in all adapter PCRs. No significant differences were found between raw errors. For the consensus10 error, all polymerases showed significantly lower error than Platinum Taq (Wilcoxon signed-rank test, p < 0.001). In addition, both Phusion and Platinum SuperFi polymerases had lower error rates than both Accuprime polymerases but were not significantly different from each other (p > 0.05). Each dot denotes a nucleotide in one of the 3 amplicons, Mean ± SD is shown (n = 3). (d) Total percentage of consensus10 reads with 0, 1, 2 or 3 variant bases. HF, High Fidelity; SF, SuperFi.