Abstract
Broad use of CRISPR-Cas12a (formerly Cpf1) nucleases1 has been hindered by the requirement for an extended TTTV protospacer adjacent motif (PAM)2. To address this limitation, we engineered an enhanced Acidaminococcus sp. Cas12a variant (enAsCas12a) that has a substantially expanded targeting range, enabling targeting of many previously inaccessible PAMs. On average, enAsCas12a exhibits two-fold higher genome editing activity on sites with canonical TTTV PAMs compared to wild-type AsCas12a, and we successfully grafted a subset of mutations from enAsCas12a onto other previously described AsCas12a variants3 to enhance their activities. enAsCas12a improves the efficiency of multiplex gene editing, endogenous gene activation, and C-to-T base editing, and we engineered a high-fidelity version of enAsCas12a (enAsCas12a-HF1) to reduce off-target effects. Both enAsCas12a and enAsCas12a-HF1 function in HEK293T and primary human T cells when delivered as ribonucleoprotein (RNP) complexes. Collectively, enAsCas12a provides an optimized version of Cas12a that should enable wider application of Cas12a enzymes for gene and epigenetic editing. [AU: Revised abstract OK?]
CRISPR-Cas nucleases are widely used for gene, epigenetic, and base editing in human cells and other organisms4-7. The study of alternative CRISPR nucleases beyond the commonly used Streptococcus pyogenes Cas9 (SpCas9), including Cas12a orthologs1,8,9, has yielded additional enzymes with distinct and potentially advantageous properties. Cas12a nucleases, including AsCas12a and Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a), recognize target sites with T-rich protospacer adjacent motifs (PAMs)1,2, require a only single short ~40 nt CRISPR RNA (crRNA) to program target specificity10, and possess ribonuclease activity that enables multiplex targeting through poly-crRNA transcript processing11. Although Cas12a enzymes have shown utility for multiplex gene editing12, gene activation13,14, and combinatorial library screens15, one constraint is their requirement for a longer PAM of the form 5’-TTTV (where V is A, C, or G), which restricts targeting approximately six-fold relative to SpCas9. Although Cas12a orthologs from Francisella novicida (FnCas12a) and Moraxella bovoculi 237 (MbCas12a) were previously reported to recognize an increased number of PAMs in vitro1, our own findings (Supplementary Figs. 1a-b and see Supplementary Results) and those previously reported by others16 have shown their activities to be less consistent and robust in human cells. Additionally, though two engineered AsCas12a variants (referred to RVR and RR) were previously described that can recognize alternative TATV and TYCV PAMs3, respectively, many PAMs remain inaccessible to Cas12a. Thus, additional variants with expanded targeting capabilities are needed to enable applications requiring high targeting density and flexibility.
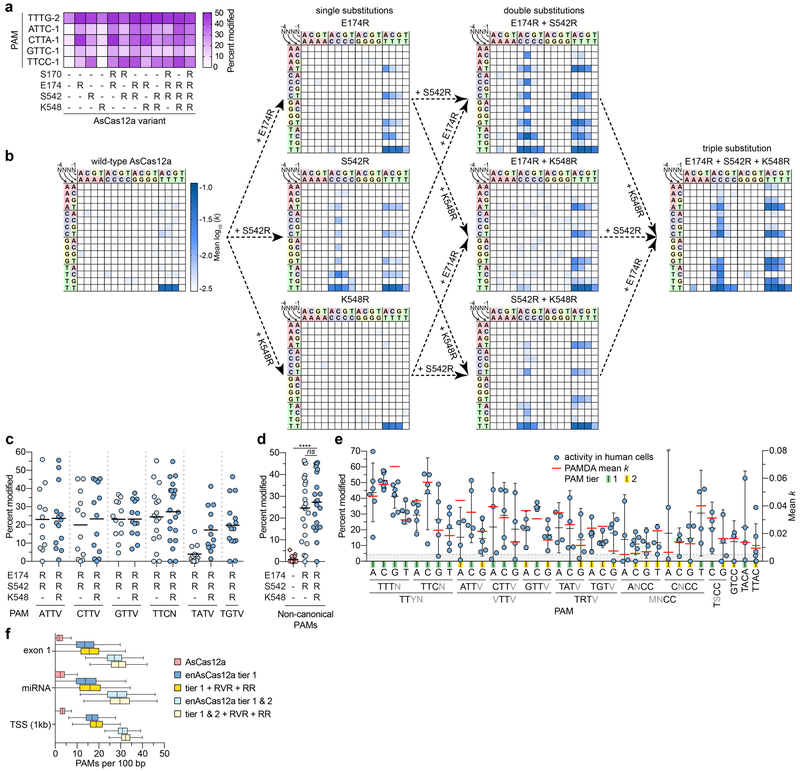
We used structure-guided protein engineering to attempt to expand the PAM recognition of Cas12a nucleases, focusing on AsCas12a because it has been widely used for genome editing and structural information was available17,18. To do so, we engineered ten variants bearing single amino acid substitutions to positively charged arginine residues that might be expected to alter or form novel PAM proximal DNA contacts (Supplementary Fig. 1c). Four of the 10 variants we tested in human cells (S170R, E174R, S542R, or K548R) displayed higher gene editing activities on sites with canonical and non-canonical PAMs relative to wild-type AsCas12a (Supplementary Figs. 1d and 1e, respectively). Testing of additional variants harboring combinations of these four substitutions on an expanded number of targets showed that two variants (E174R/S542R and E174R/S542R/K548R) exhibited the highest editing activities on sites with non-canonical PAMs while still retaining robust activities on a canonical PAM site (Fig. 1a and Supplementary Fig. 1f).
Figure 1. Engineering and characterization of AsCas12a variants with expanded target range in human cells.
(a) Modification of endogenous sites in human cells by AsCas12a variants, assessed by T7E1 assay; mean shown for n ≥ 3. (b) PAM preference profiles, assessed by PAMDA, for wild-type AsCas12a and all intermediate single and double substitution variants that comprise the E174R/S542R/K548R variant. The log10 rate constants (k) are the mean of four replicates, two each against two distinct spacer sequences (see Supplementary Fig. 2d). (c) Mean activity plots for AsCas12a variants on sites with non-canonical PAMs in human cells, where the black line represents the mean of 12 to 20 sites (dots) for each PAM class (see also Supplementary Figs. 3a, 3b and 3e). (d) Summary of the activities of wild-type AsCas12a and variants across sites in human cells encoding non-canonical PAMs, one for each PAM of the VTTN, TTCN, TATN, and TTTT classes (from Supplementary Figs. 1a, 3a-3c all sites numbered ‘1’, and all sites in Supplementary Fig. 3d); mean shown for n = 20; ns, P > 0.05; ****, P < 0.001 (Wilcoxon signed-rank, two-tailed; P values in Supplementary Table 8). (e) Superimposition of the summaries of the human cell activities and PAMDA rate constants (k) for various targetable PAMs with enAsCas12a (E174R/S542R/K548R); mean and 95% confidence interval for human cell data shown with black lines. Tier 1 PAMs exhibit greater than 20% mean targeting in human cells and a PAMDA k greater than 0.01; tier 2 PAMs meet a modest threshold of greater than 10% mean targeting in cells and a PAMDA k greater than 0.005 (see Supplementary Table 2). (f) Calculation of the improvements in targeting range enabled by AsCas12a variants compared to wild-type AsCas12a, plotted as the number of PAMs per 100 bp window as determined by enumerating complete PAM sequences within the indicated sequence feature and normalizing for element size (see Methods). TSS, transcription start site; PAM sequences targetable by wild-type AsCas12a, TTTV; by RVR, TATV; and by RR, TYCV.
To comprehensively profile the PAM preferences of these AsCas12a variants, we optimized an unbiased in vitro PAM determination assay (PAMDA) similar to other previously described methods3,19 (see Supplementary Results, Supplementary Figs. 2a-2i, and Supplementary Table 1). Using the PAMDA, we defined the PAM preferences of wild-type AsCas12a and variants with all possible single, double, and triple combinations of the E174R, S542R, and K548R substitutions. Plots of the mean PAMDA log10k values on all 256 4-nt PAM sequences revealed that, as expected, targeting with wild-type AsCas12a was most efficient against TTTV PAMs, and that E174R/S542R and E174R/S542R/K548R showed the most expanded PAM preferences among the seven variants tested (Fig. 1b). Strikingly, the E174R/S542R/K548R variant could target many PAMs including TTYN (TTTN/TTCN), VTTV (ATTV/CTTV/GTTV), TRTV (TATV/TGTV), and others.
Further characterization of the E174R/S542R and E174R/S542R/K548R variants in human cells showed robust editing activities on 60 endogenous target sites with VTTV and TTCN PAMs, and less effective modification of 15 target sites with VTTT PAMs (Fig. 1c and Supplementary Fig. 3a). Consistent with the PAMDA results (Fig. 1b), we observed efficient gene editing on 12 target sites bearing TATV PAMs with the E174R/S542R/K548R variant but not with E174R/S524R (Fig. 1c and Supplementary Fig. 3b). Both variants modified five sites with TTTT PAMs that were inefficiently edited with wild-type AsCas12a (Supplementary Fig. 3c). These data show that our variants enable robust editing of sites with non-canonical VTTV, TTTT, and TTCN/TATV PAMs that cannot be modified efficiently by wild-type AsCas12a (summarized in Fig. 1d; see also Supplementary Figs. 1a, 3a-3d).
Next, we examined the editing activities of the E174R/S542R/K548R variant on 97 other sites in human cells bearing 28 additional PAMs identified as targetable by the PAMDA. We observed efficient modification of 14 of 15 sites with TGTV PAMs (Supplementary Fig. 3e) and a range of editing activities across the other 82 sites harboring 25 additional PAMs (Supplementary Fig. 3f). Comparison of the mean PAMDA log10k and the mean human cell targeting values across the same PAMs showed a strong correlation for most PAMs (Supplementary Fig. 3g), suggesting that PAMs with a PAMDA log10k of −2.25 or higher were potentially targetable in human cells. PAMs accessible with E174R/S542R/K548R were binned into confidence tiers based on consistency between PAMDA and human cell experiments (Fig. 1e, Supplementary Fig. 3h, and Supplementary Table 2; see Supplementary Results). Differences in activities across sites with the E174R/S542R/K548R variant could not be attributed to PAM or spacer sequence features (Supplementary Figs. 3i-3n; see Supplementary Results). Taken together, our combined analyses illustrate that the E174R/S542R/K548R variant, henceforth referred to as enhanced AsCas12a (enAsCas12a), expands targeting by approximately seven-fold (Fig. 1f).
While characterizing our different engineered AsCas12a variants, we noticed that certain substitutions were associated with increased editing activities in human cells (Fig. 1a and Supplementary Fig. 1d). To assess this improvement more comprehensively, we compared the gene modification activities of wild-type AsCas12a, E174R/S542R, and enAsCas12a across 21 endogenous gene sites with canonical PAMs (Supplementary Fig. 4a). Compared to wild-type AsCas12a, both variants were on average nearly two-fold more effective at modifying sites with TTTV PAMs (Fig. 2a).
Figure 2. AsCas12a variants enhance on-target editing in human cells.
(a) Mean activity plots for AsCas12a, E174R/S542R, and enAsCas12a on sites with TTTV PAMs, where the black line represents the mean of 21 sites (see also Supplementary Fig. 4a); ns, P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Wilcoxon signed-rank, two-tailed; P values in Supplementary Table 8). (b) Quantification of time-course in vitro cleavage reactions of Cas12a orthologs and variants on linearized plasmid substrates encoding the PAMDA site 1 target, conducted at 37, 32, and 25 °C (left, middle, and right panels, respectively). Curves were fit using a one phase exponential decay equation; mean and error bars represent s.e.m for n = 3. (c-e) Summaries of the activities of wild-type AsCas12a and variants across sites encoding TTTN PAMs (panel c; n = 11), TATN PAMs (panel d; n = 14) and TYCN PAMs (panel e; n = 29) (see also Supplementary Figs. 4b-4d, respectively); mean activity shown with black line; ns, P > 0.05; *, P < 0.05; **, P < 0.001 (Wilcoxon signed-rank, two-tailed; P values in Supplementary Table 8). (f) Scatterplots of the PAMDA determined rate constants for each NNNN PAM to compare the PAM preferences of AsCas12a variants (RVR to enRVR, left panel; RR to enRR, right panel). Variants encode the following substitutions: enAsCas12a, E174R/S542R/K548R; RVR, S542R/K548V/N552R; enRVR, E174R/S542R/K548V/N552R; RR, S542R/K607R; enRR, E174R/S542R/K607R.
Given the enhanced gene editing efficiencies observed with enAsCas12a, we speculated that this variant might also exhibit improved activity at lower temperatures, a property relevant for organisms that grow optimally at temperatures lower than 37 °C. Previous studies showed that editing with AsCas12a had decreased activity at lower temperatures relative to LbCas12a20-22. In vitro cleavage reactions of AsCas12a, LbCas12a, and enAsCas12a at 37, 32, and 25 °C revealed that enAsCas12a is more active than AsCas12a at these temperatures, exhibiting activities more comparable to LbCas12a (Fig. 2b). Systematic examination of variants harboring all possible combinations of the E174R, S542R, and K548R substitutions revealed that the improvements in cleavage efficiency with enAsCas12a at lower temperatures were largely attributable to E174R and to a lesser extent to S542R (Fig. 2b).
To determine whether the increased activity phenotype of enAsCas12a might be transferable to other AsCas12a variants, we added the E174R substitution to the previously described RVR and RR PAM recognition variants3 to create enRVR and enRR, respectively. Comparison of the activities of wild-type AsCas12a, enAsCas12a, RVR, enRVR, RR, and enRR across 11 sites with TTTN PAMs in human cells revealed that the original RVR and RR variants have similar or weaker activities compared to wild-type AsCas12a (Supplementary Fig. 4b). Furthermore, the enRVR and enRR variants generally showed more than two-fold higher activities than RVR and RR, albeit with lower activities compared to enAsCas12a (Fig. 2c). Comparison of the editing activities of RVR and enRVR on 14 endogenous gene sites bearing TATN PAMs (Supplementary Fig. 4c) again showed an approximately two-fold improvement with enRVR relative to the original RVR variant (Fig. 2d). Similarly, we examined the activities of enRR and RR in human cells on 29 sites bearing TYCN PAMs (Supplementary Fig. 4d) and found that enRR showed an average of 1.5-fold improved efficiency compared to RR (Fig. 2e). Importantly, the enRVR and enRR variants retained similar PAM preferences to their parental RVR and RR variants (Fig. 2f and Supplementary Fig. 4e). Collectively, enAsCas12, enRVR, and enRR exhibit improved activities and can target an expanded range of sequences compared wild-type AsCas12a (Supplementary Fig. 4f and Supplementary Table 2).
We next sought to assess whether enAsCas12a could improve the efficiency of various Cas12a-based applications. One potential advantage of Cas12a enzymes is their ribonuclease activity, which enables the processing of individual crRNAs from a poly-crRNA transcript11 and simplifies multiplex targeting in cells12-14. We compared the activities of AsCas12a, enAsCas12a, and LbCas12a programmed with poly-crRNA arrays targeted to three endogenous genes in human cells (Figs. 3a-3c). In all cases, we observed comparable or higher editing with enAsCas12a relative to AsCas12a and LbCas12a. We also designed multiplex arrays encoding two sets of proximally targeted crRNAs to generate small genomic deletions. Pairs of crRNAs were expressed from either poly-crRNA transcripts or pools of single crRNA plasmids, and we again observed comparable or improved deletion efficiencies with enAsCas12a relative to AsCas12a and LbCas12a (Fig. 3d).
Figure 3. Improved multiplex editing, gene activation, and base editing with enAsCas12a.
(a-c) Comparison of the multiplex modification efficiencies of AsCas12a, enAsCas12a, and LbCas12a, when programmed with TTTV PAM targeted crRNA arrays encoding 3 separate crRNAs expressed either from a polymerase III promoter (U6, panels a and b) or a polymerase II promoter (CAG, panel c). The activities at three separate loci were assessed by T7E1 assay using the same genomic DNA samples; mean, s.e.m., and individual data points shown for n = 3. (d) Assessment of editing efficiencies with AsCas12a, enAsCas12a, and LbCas12a when using pooled crRNA plasmids or multiplex crRNA arrays expressing two crRNAs targeted to nearby (~100 bp) genomic loci. Activities assessed by T7E1 assay; mean, s.e.m., and individual data points shown for n = 4. (e-g), Activation of endogenous human genes NPY1R, HBB, and AR with dCas12a-VPR(1.1) fusions in HEK293 cells using pools of three crRNAs targeted to canonical PAM sites (panel e) and non-canonical PAM sites (panels f and g). Activities assessed by RT-qPCR and fold-changes in RNA were normalized to HPRT1 levels; mean, s.e.m., and individual data points shown for three independent experiments (mean of three technical triplicate qPCR values); VPR, synthetic VP64-p65-Rta activation domain26. (h) Cytosine to thymine (C-to-T) conversion efficiencies directed by dCas12a base-editor (BE) constructs across eight different target sites, assessed by targeted deep sequencing. The mean percent C-to-T editing of three independent experiments was examined within a −5 to +25 window; all Cs in this window are highlighted in green for each target site; the position of the C within the target site is indicated below the heat map. (i) C-to-T editing efficiency within the 20 nt target site spacer sequence with enAsBEs and LbBEs across all eight target sites.
Cas12a has also been used for epigenetic editing of endogenous human and plant genes by fusing DNase-inactive Cas12a (dCas12a) to heterologous effectors13,21,23-25. We found that a fusion of DNase-inactive enAsCas12a fusion to the synthetic VPR activation domain26 (denAsCas12a-VPR) outperformed analogous dAsCas12a-, dLbCas12a-13, and dSpCas926-based VPR fusions (Supplementary Results and Supplementary Figs. 5a-5f). Additional experiments comparing dAsCas12a-, denAsCas12a-, and dLbCas12a-VPR fusions targeted to the promoters of three endogenous human genes revealed that the most potent gene activation (range of 10- to 10,000-fold upregulation) was consistently achieved with the denAsCas12a-VPR fusion using pools of crRNAs targeted to sites with canonical (Fig. 3e) or non-canonical PAMs (Figs. 3f and 3g).
Cas12a enzymes have recently been adapted for base editing to induce targeted C-to-T alterations27. Base editors (BEs) consist of cytosine deaminases and uracil glycosylase inhibitor (UGI) domains fused to nickase versions of Cas9 or DNase-inactive forms of LbCas12a27-30. Comparable DNase-inactive AsCas12a-BEs (AsBEs) have been reported as being minimally active27. To determine whether the enhanced activities of enAsCas12a could enable efficient base editing, we compared four different denAsCas12a base editor fusions (enAsBE1.1-1.4; Supplementary Fig. 6a) to two analogous dAsCas12a constructs (AsBE1.1 and 1.4). Consistent with previous reports27, we observed minimal (< 2%) C-to-T editing with AsBEs across all Cs for seven of eight sites in human cells with a maximum of 6% editing on the eighth site (Fig. 3h and Supplementary Table 3). However, enAsBE fusions exhibited substantially improved C-to-T editing across the same eight sites (range of 2-34% editing; Fig. 3h and Supplementary Fig. 6b). Assessment of two analogous dLbCas12a base editors (LbBE1.1 and 1.4) revealed levels of C-to-T editing comparable to those of enAsBEs (range of 2-19% C-to-T editing; Figs. 3h and 3i). Similar to previous studies of SpCas9-BEs, GC motifs were edited less efficiently by Cas12a-BEs than AC, CC or TC motifs (Supplementary Fig. 6c). Additionally, C-to-T conversion was the predominant edit outcome with enAsBEs and LbBEs (Supplementary Fig. 6d), while levels of insertion and deletion mutations (indels) for Cas12a-BEs were low presumably due to DNase inactivation of the nuclease. (Supplementary Fig. 6e).
Because enAsCas12a targets an expanded number of PAMs, we assessed the genome-wide specificity of this variant in human cells using GUIDE-seq31 (Supplementary Fig. 7a). Experiments performed with four crRNAs targeted to sites harboring TTTV PAMs (Supplementary Figs. 7b-7d) showed few detectable off-targets with wild-type AsCas12a by GUIDE-seq but additional off-targets with enAsCas12a (Fig. 4a, Supplementary Fig. 7e, and Supplementary Table 4). Several of the off-targets observed only with enAsCas12a were sites that harbored non-canonical PAMs, were previously identified as off-target sites in GUIDE-seq experiments performed with LbCas12a and the same crRNAs10, or contained mismatches in spacer positions known to be tolerant of nucleotide substitutions by AsCas12a10,16,32.
Figure 4. Characterization and improvement of AsCas12a specificity and activity.
(a, b) Histograms illustrating the number of GUIDE-seq detected off-target sites for AsCas12a variants on sites with canonical TTTV PAMs (panel a; see Supplementary Fig. 7e) or non-canonical PAMs (panel b; see Supplementary Fig. 7f). na, not assessed. (c, d) Summaries of the on-target activities of wild-type, enAsCas12a, and enAsCas12a-HF1 across sites encoding TTTV PAMs (panel c; n = 6) or enAsCas12a and enAsCas12a-HF1 on non-canonical PAMs (panel d; n = 17) (see Supplementary Figs. 9b and 9c, respectively). (e) Assessment of the gene editing activities of AsCas12a, enAsCas12a, and enAsCas12a-HF1 on target sites harboring TTTV PAMs or non-canonical PAMs (n = 5 and 6, respectively) in primary human T cells when delivered as RNPs (see Supplementary Fig. 9f). For panels c-e, percent modified assessed by T7E1 assay, mean shown by black bar, and each point is the mean of 3 independent experiments (see Supplementary Figs. 9b, 9c, and 9f); ns, P > 0.05; *, P < 0.05 (Wilcoxon signed-rank, two-tailed; P values in Supplementary Table 8). Variants encode the following substitutions: enAsCas12a, E174R/S542R/K548R; enAsCas12a-HF1, E174R/N282A/S542R/K548R.
To attempt to improve the specificity of enAsCas12a, we employed a strategy that we and others previously employed to engineer high-fidelity variants of SpCas933-35. Using structure-guided design, we created a series of AsCas12a and enAsCas12a variants with substitutions in amino acid residues expected to make non-specific contacts to DNA17 (see Supplementary Results and Supplementary Figs. 8a and 8b). Among the variants we examined, we found that enAsCas12a-N282A exhibited the greatest improvement in single mismatch intolerance while retaining on-target activity similar to enAsCas12a (Supplementary Fig. 8b). Comparison of the two nucleases using the PAMDA revealed nearly identical PAM preference profiles (Supplementary Figs. 8c-8e), suggesting that the N282A substitution does not substantially alter targeting range.
We compared the genome-wide specificities of enAsCas12a and enAsCas12a-N282A using GUIDE-seq performed with the same four TTTV PAM crRNAs described above. The introduction of the N282A substitution into enAsCas12a reduced both the number of off-target sites and the magnitude of GUIDE-seq read counts at off-target sites for three of the four crRNAs (Fig. 4a and Supplementary Fig. 7e). Additional GUIDE-seq experiments using 10 crRNAs targeted to sites with non-canonical PAMs again revealed that enAsCas12a-N282A reduced the number of off-target sites and GUIDE-seq read counts compared to enAsCas12a (Fig. 4b and Supplementary Fig. 7f). Based on these results, we conclude that the N282A substitution can be combined with enAsCas12a to generate an enhanced high-fidelity AsCas12a variant, which we refer to as enAsCas12a-HF1.
To more thoroughly determine if the N282A substitution impacts on-target activity, we compared enAsCas12a and enAsCas12a-HF1 using several methods. We first performed in vitro cleavage assays to assess temperature tolerance, which revealed similar cleavage profiles among enAsCas12a, enAsCas12a-HF1, and LbCas12a at 37, 32, and 25 °C (Supplementary Fig. 9a). Next, we compared the on-target activities of both AsCas12a variants when delivered by plasmid electroporation into human U2OS cells on sites with canonical and non-canonical PAMs (Supplementary Figs. 9b and 9c, respectively). These experiments revealed similar on-target activities for the enAsCas12a and enAsCas12a-HF1across six sites with TTTV PAMs, where both variants again exhibited ~2-fold improved editing compared to wild-type AsCas12a (Fig. 4c). Additionally, we observed comparable activities of enAsCas12a and enAsCas12a-HF1 on 17 target sites with various non-canonical PAMs (Fig. 4d). Taken together, these results show that introduction of N282A does not abrogate the improved targeting range, temperature tolerance, or higher gene editing activities of enAsCas12a-HF1.
Because the delivery of nucleases as ribonucleoprotein (RNP) complexes offers advantages for research use and potentially therapeutic applications36-38, we assessed whether our enhanced variants could be transfected as RNPs into HEK293T and primary human T cells. We performed initial experiments with Cas12a RNPs in HEK293T cells to compare gene disruption efficiencies of wild-type AsCas12a, enAsCas12a, and enAsCas12-HF1 targeted to sites with canonical and non-canonical PAMs (see Supplementary Results and Supplementary Figs. 9d and 9e). We then examined the activities of the same RNP complexes when delivered to primary human T cells (Supplementary Fig. 9f) and found that enAsCas12a and enAsCas12a-HF1 both showed a nearly two-fold mean improvement in on-target editing on sites with TTTV PAMs compared to wild-type AsCas12a (Fig. 4e). Furthermore, enAsCas12a and enAsCas12a-HF1 also exhibited editing on sites with non-canonical PAMs compared with negligible editing by wild-type AsCas12a on these same sites (Fig. 4e).
The enhanced AsCas12a variants described herein substantially improve the targeting range, on-target activities, and fidelity of Cas12a nucleases, properties that are important for multiplex gene editing, epigenetic editing, cytosine base editing, and gene knockout in primary human T cells. Our in vitro PAMDA and human cell experiments suggest that enAsCas12a can target approximately 1 in every 6 bps of DNA, a roughly seven-fold improvement compared to most Cas12a orthologs. enAsCas12a also exhibits superior on-target activity relative to wild-type AsCas12a, increasing editing efficiencies by approximately two-fold on sites with canonical TTTV PAMs in two cell lines and in primary human T cells. The enhanced enRVR and enRR variants also show improved activities compared to their parental variants. Our results provide an important proof-of-concept that the on-target potency of CRISPR enzymes can be augmented through engineering, a strategy that may be extensible to other CRISPR nucleases. Future structural studies will be helpful to characterize the roles of the substitutions in our AsCas12a variants (see Supplementary Discussion), and additional work may be required to determine whether the potency of enAsCas12a and enAsCas12a-HF1 RNPs are sufficient for therapeutic applications. In sum, the superior properties of the enhanced Cas12a enzymes described herein enable a wide range of applications that should encourage more widespread adoption of this class of nucleases.
Online Methods
Plasmids and oligonucleotides.
New plasmids described in this study have been deposited with the non-profit plasmid repository Addgene (http://www.addgene.org/crispr-cas)). Descriptions and sequences of plasmids can be found in Supplementary Table 5 and the Supplementary Information, respectively. The target site sequences for crRNAs and oligonucleotide sequences are available in Supplementary Tables 6 and 7, respectively. Human expression plasmids for wild-type AsCas12a, LbCas12a, FnCas12a, and MbCas12a (SQT1659, SQT1665, AAS1472, AAS2134, respectively) were generated by sub-cloning the nuclease open-reading frames from plasmids pY010, pY016, pY004, and pY014, respectively (Addgene plasmids 69982, 69988, 69976, and 69986; gifts from Feng Zhang) into the NotI and AgeI sites of pCAG-CFP (Addgene plasmid 11179; a gift from Connie Cepko). Protein expression plasmids were generated by cloning the human codon-optimized open reading frame of AsCas12a and the bacterial codon-optimized LbCas12a open reading frame (from Addgene plasmid 79008; a gift from Jin Soo Kim) into the NcoI and FseI sites of pET28b-Cas9 (Addgene plasmid 47327; a gift from Alex Schier) to generate BPK3541 and RTW645, respectively. All Cas12a variants, activator constructs, and base-editor fusions were generated via standard molecular cloning and isothermal assembly. Human cell expression plasmids for Cas12a crRNAs were generated by annealing and ligating oligonucleotides corresponding to spacer sequence duplexes into BsmBI-digested BPK3079, BPK3082 (ref. 10), BPK4446, and BPK4449 for U6 promoter-driven transcription of As, Lb, Fn, and MbCas12a crRNAs, respectively. Substrate plasmids for in vitro cleavage reactions were generated by cloning target sites into the EcoRI and SphI sites of p11-lacY-wtx1. Plasmids for in vitro transcription of Cas12a crRNAs were generated by annealing and ligating oligonucleotides corresponding to spacer sequence duplexes into BsaI-digested MSP3491 and MSP3495 for T7 promoter-driven transcription of As and LbCas12a crRNAs, respectively.
Cell culture conditions and isolation of primary human T cells.
Human U2OS (from Toni Cathomen, Freiburg) were cultured in Advanced Dulbecco’s Modified Eagle Medium (A-DMEM) supplemented with 10% heat-inactivated FBS (HI-FBS), 1% penicillin/streptomycin, and 2 mM GlutaMax. HEK293 cells (Invitrogen) and HEK293T (ATCC) cells were cultured in DMEM supplemented with 10% HI-FBS and 1% penicillin/streptomycin (with the exceptions that HEK293 cells cultured for experiments analyzed by RT-qPCR had 0.4% penicillin/streptomycin and HEK293 cells cultured for experiments analyzed by ELISA were also supplemented with 2 mM GlutaMax). Primary human T cells were cultured in RPMI1640 supplemented with 10% HI-FBS, 1% penicillin/streptomycin, 1% GlutaMax, 1% non-essential amino acids, 1% sodium pyruvate, 5mM HEPES, 50 μM 2-mercaptoethanol (Millipore-Sigma), and 20 IU/mL IL-2 (Peprotech). 1.5% M form Phytohemagglutinin were added to T Cell cultures upon thaw. Cell culture reagents were purchased from Thermo Fisher Scientific unless otherwise noted, and cells were grown at 37 °C in 5% CO2. Media supernatant was analyzed biweekly for the presence of Mycoplasma using MycoAlert PLUS (Lonza), and cell line identities were confirmed by STR profiling (ATCC). No commonly misidentified cell lines were used. Unless otherwise indicated, negative control transfections included Cas12a and U6-null plasmids.
Primary Human T cells were isolated from Source Leukocytes purchased from the Massachusetts General Hospital Blood Transfusion Service with prior approval from the Partners Subcommittee on Human Studies. T cells were enriched from whole blood using RosetteSep Human T Cell Enrichment Cocktail (STEMCELL Technologies) per manufacturer instructions. Following T cell enrichment, 15mL of RosetteSep-treated blood was mixed 1:1 with PBS containing 2% HI-FBS and then gently overlaid onto 15 mL of Ficoll-Paque Plus (Millipore-Sigma). Phase separated suspensions were centrifuged at 1200 g for 20 minutes with no brake applied. Following centrifugation, the buffy coat fraction was removed and washed twice with 45mL of PBS supplemented with 2% HI-FBS. Cells were centrifuged at 300 g for 10 minutes with minimum brake following each wash. Washed T cells were resuspended in in 90% HI-FBS and 10% DMSO (ATCC) and aliquots of approximately 40 million cells were cryopreserved.
Assessment of gene and base editing by T7E1 or deep-sequencing.
For nuclease and base-editor experiments in U2OS cells, Cas12a and crRNA expression plasmids (580 ng and 250 ng, respectively) were electroporated into approximately 2×105 U2OS cells via the DN-100 program with the SE Cell Line Nucleofector Kit using a 4D-Nucleofector (Lonza). For ribonucleoprotein (RNP) experiments in HEK293T cells, approximately 105 cells were seeded in a 24-well plate about 24 hours prior to transfection. RNPs were complexed by mixing 70 pmol of Cas12a and 140 pmol crRNA (Integrated DNA Technologies) in 50 uL Opti-MEM at room temperature for 15 minutes. Next, RNPs were mixed with with 4 μL CRISPRMAX and 2.5 μL Cas9 Plus reagent (Invitrogen), and then carefully dropped into existing cell culture media for transfection. For experiments in T cells, after two days in culture, Cas12a RNPs (70 pmol of Cas12a and 140 pmol crRNA) were electroporated into approximately 3×105 cells via the DN-100 program with the P3 Primary Cell Nucleofector Kit using a 4D-Nucleofector.
Genomic DNA (gDNA) was extracted approximately 72 or 120 hours post-electroporation (for nuclease or base editing experiments, respectively) using the Agencourt DNAdvance Nucleic Acid Isolation Kit (Beckman Coulter), or by custom lysis and paramagnetic bead extraction. Paramagnetic beads were prepared similar to as previously described39 (GE Healthcare Sera-Mag SpeedBeads from Fisher Scientific, washed in 0.1x TE and suspended in 20% PEG-8000 (w/v), 1.5 M NaCl, 10 mM Tris-HCl pH 8, 1 mM EDTA pH 8, and 0.05% Tween20). For cell lysis, media supernatant was removed, a 500 μL PBS wash was performed, and the cells were treated with 200 μL lysis buffer (100 mM Tris HCl pH 8.0, 200 mM NaCl, 5 mM EDTA, 0.05% SDS, 1.4 mg/mL Proteinase K (New England Biolabs; NEB), and 12.5 mM DTT) for 12-20 hours at 55 °C. To extract gDNA, the lysate was combined with 165 μL paramagnetic beads, mixed thoroughly, incubated for 5 minutes, separated on a magnetic plate and washed 3 times with 70% EtOH, allowed to dry for 5 minutes, and eluted in 65 μL elution buffer (1.2 mM Tris-HCl pH 8.0). Genomic loci were amplified by PCR with Phusion Hot Start Flex DNA Polymerase (NEB) using approximately 100 ng of gDNA as a template and the primers listed in Supplementary Table 7. Following analysis on a QIAxcel capillary electrophoresis machine (Qiagen), PCR products were purified with paramagnetic beads.
For nuclease experiments, the percent modification of endogenous human target sites was determined by T7 Endonuclease I (T7EI) assays, similar to as previously described40. The T7E1 assay was selected to quantify relative the differences in activities between Cas12a nucleases because it has previously been shown that the T7E1 assay is effective at detecting indels greater than 1 nt 41,42, consistent with indel profiles that are commonly observed with Cas12a nucleases16,43. Briefly, 200 ng of purified PCR products were denatured, annealed, and digested with 10 U T7EI (NEB) at 37 °C for 25 minutes. Digests were purified with paramagnetic beads and analyzed using a QIAxcel to estimate target site modification.
For base editing experiments, targeted deep sequencing was performed essentially as previously described33. Dual-indexed Tru-seq libraries were generated from purified and pooled PCR products using a KAPA HTP Library Preparation Kit (KAPA BioSystems) and sequenced on an Illumina MiSeq Sequencer. Samples were sequenced to an average of 57,833 reads (minimum of 8,534 reads) per replicate and an average of 173,499 (minimum of 70,022) per triplicate condition. Nucleotide substitutions and insertion or deletion mutations (indels) were analyzed using CRISPResso244 (Supplementary Table 3), with an additional custom analysis performed to examine indel percentages (defined as [reads with an indel and/or substitution – substitution only reads] / total reads *100), in a 44 nt window encompassing the −14 to +30 region of each target site (an additional 10 nt upstream of the 4 nt PAM and 10 nt downstream of the 20 nt spacer sequence).
Gene activation experiments.
For experiments with crRNAs or sgRNAs targeting the VEGFA promoter, 1.6×105 HEK293 cells per well were seeded in 24-well plates roughly 24 hours prior to transfection with plasmids encoding Cas12a or Cas9 activators and pools of crRNAs or sgRNAs (750 ng and 250 ng, respectively), 1.5 μL TransIT-LT1 (Mirus), and Opti-MEM to a total volume of 50 μL. The cell culture media was changed 22 hours post-transfection, and aliquots of the media supernatant were collected 44 hours post-transfection to determine VEGFA concentration using a Human VEGF Quantikine ELISA Kit (R&D Systems).
For experiments with crRNAs targeting the AR, HBB, or NPY1R promoters, 8.6×104 HEK293 cells per well were seeded in 12-well plates roughly 24 hours prior to transfection with 750 ng Cas12a activator expression plasmid, 250 ng crRNA plasmid pools, 3 μL TransIT-LT1 (Mirus), and 100 μL Opti-MEM. Total RNA was extracted from the transfected cells 72 hours post-transfection using the NucleoSpin RNA Plus Kit (Clontech). cDNA synthesis using a High-Capacity RNA-to-cDNA kit (ThermoFisher) was performed with 250 ng of purified RNA, and 3 μL of 1:20 diluted cDNA was amplified by quantitative reverse transcription PCR (RT-qPCR) using Fast SYBR Green Master Mix (ThermoFisher) and the primers listed in Supplementary Table 7. RT-qPCR reactions were performed on a LightCycler480 (Roche) with the following cycling program: initial denaturation at 95 °C for 20 seconds (s) followed by 45 cycles of 95 °C for 3 s and 60 °C for 30 s. If sample amplification did not reach the detection threshold after 35 cycles, Ct (Cycles to threshold) values were considered as 35 due to Ct fluctuations typical of transcripts expressed at very low levels. Gene expression levels over negative controls experiments (Cas12a activator and empty crRNA plasmids) were normalized to the expression of HPRT1.
GUIDE-seq.
GUIDE-seq experiments were performed as previously described. Briefly, U2OS cells were electroporated as described above but including 100 pmol of the double-stranded oligodeoxynucleotide (dsODN) GUIDE-seq tag. Restriction-fragment length polymorphisms (RFLP) assays (performed as previously described45) and T7E1 assays (as described above) were performed to assess GUIDE-seq tag integration and on-target modification percentages, respectively. GUIDE-seq libraries were sequenced using an Illumina MiSeq sequencer, and data was analyzed using guideseq v1.1 (ref. 46) with an NNNN PAM, a 75 bp window, and allowing up to 9 mismatches prior to downstream data processing (Supplementary Table 4). High-confidence, cell-type-specific, single-nucleotide polymorphisms (SNPs) were identified using SAMTools.
Expression and purification of Cas12a proteins.
For in vitro experiments: Plasmids encoding Cas12a-NLS(nucleoplasmin)-6xHis fusion proteins were transformed into Rosetta 2 (DE3) E. coli and single colonies were inoculated into 25 mL LB medium cultures containing 50 mg/L kanamycin and 25 mg/L chloramphenicol (Kan/Cm) prior to growth at 25 °C for 16 hours. Starter cultures were then diluted 1:100 into 150 mL LB medium containing Kan/Cm and grown at 37 °C until the OD600 reached 0.4. Cultures were then induced with 0.2 mM isopropyl β-D-thiogalactopyranoside prior to shaking at 18 °C for 23 hours. Cell pellets from 50 mL of culture were harvested by centrifugation at 1200 g for 15 minutes and suspended in 1 mL lysis buffer v1 containing 20 mM Hepes pH 7.5, 100 mM KCl, 5 mM MgCl2, 5% glycerol, 1 mM DTT, Sigmafast protease inhibitor (Sigma-Aldrich), and 0.1% Triton X-100. The cell suspension was loaded into a 1 mL AFA fiber milliTUBE (Covaris) and was lysed using an E220evolution focused-ultrasonicator (Covaris) according to the following conditions: peak intensity power of 150 W, 200 cycles per burst, duty factor of 10%, and treatment for 20 minutes at 5 °C. The cell lysate was centrifuged for 20 minutes at 21,000 g and 4 °C, and the supernatant was mixed with an equal volume of binding buffer v1 (lysis buffer v1 with 10 mM imidazole), added to 400 μL of HisPur Ni-NTA Resin (Thermo Fisher Scientific) that was pre-equilibrated in binding buffer v1, and rocked at 4 °C for 8 hours. The protein-bound resin was washed three times with 1 mL wash buffer v1 (20 mM Hepes pH 7.5, 500 mM KCl, 5 mM MgCl2, 5% glycerol, 25 mM imidazole, and 0.1% Triton X-100) and then once with 1 mL binding buffer v1. Three sequential elutions were performed with 500 μL elution buffer (20 mM Hepes pH 7.5, 100 mM KCl, 5 mM MgCl2, 10% glycerol, and 500 mM imidazole) and visualized by SDS polyacrylamide gel electrophoresis and coomassie staining. Select elutions were pooled and dialyzed using Spectra/Por 4 Standard Cellulose Dialysis Tubing (Spectrum Chemical Manufacturing Corp) in three sequential 1:500 buffer exchanges, the first two into dialysis buffer (300 mM NaCl, 10 mM Tris-HCl pH 7.4, 0.1 mM EDTA, and 1 mM DTT) and the last into dialysis buffer containing 20% glycerol. Proteins were then concentrated with Amicon Ultra-0.5 mL Centrifugal Filter Units (Millipore Sigma), diluted with an equal volume of dialysis buffer with 80% glycerol to final storage conditions of 1x dialysis buffer with 50% glycerol, and stored at −20 °C.
For experiments in human cells: Starter cultures were grown as described above. After 16 hours, cultures were diluted 1:100 into ZYP-5052 auto-induction media (prepared as previously described47) containing Kan/Cm and grown at 37 °C until the OD600 reached 1.5-2. Cultures were then grown at 18 °C for an additional 24 hours. Cultures were harvested by centrifugation at 4 °C and 1200 g for 15 minutes and pellets were either stored at −80 °C or processed immediately. All subsequent steps were performed at 4 °C. Cell pellets were resuspended in 10 mL per gram cell pellet lysis buffer v2 (50 mM Tris HCL pH 7.5, 500 mM NaCl, 5% glycerol, 1 mM DTT, 0.1% Triton-X100, Sigmafast protease inhibitor) supplemented with benzonase nuclease at 2 U/mL (EMD Millipore) and lysozyme (Millipore Sigma) at 0.5 mg/mL. Bacteria were lysed with a Branson 450 sonicator for 10 minutes. The lysate was centrifuged at 21,000 g for 20 minutes and the supernatant was collected, supplemented with imidazole to 10 mM, and applied to 1.5 mL HisPur Ni-NTA Resin (Thermo Fisher Scientific) that was pre-equilibrated in binding buffer v2 (50 mM Tris-HCl pH 7.5, 500 mM NaCl, and 10 mM imidazole). The resin was rotated with the lysate for 7 hours, was washed 4 times with 10 resin-volumes of wash buffer v2 (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 25 mM imidazole, 5% glycerol, 1 mM DTT, and 0.1% Triton X-100), and then remaining bound proteins were eluted with three aliquots of 4.5 mL of elution buffer v2 (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 500 mM imidazole, 10% glycerol, and 1 mM DTT). Eluted fractions were pooled, concentrated with a 100 MWCO Amicon Ultra-15 mL Centrifugal Filter Unit (Millipore Sigma), and subjected to size exclusion chromatography in dialysis buffer on an ÄKTA FPLC (GE Healthcare) with a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences). Fractions were pooled and concentrated with a 100 MWCO Amicon Ultra-15 mL Centrifugal Filter Unit. Concentrated protein was diluted in dialysis buffer with 80% glycerol to final storage conditions of 1X dialysis buffer with 50% glycerol. Purified proteins were stored at −20 °C.
In vitro cleavage reactions.
Cas12a crRNAs were in vitro transcribed from roughly 1 μg of HindIII linearized crRNA transcription plasmid using the T7 RiboMAX Express Large Scale RNA Production kit (Promega) at 37 °C for 16 hours. The DNA template was degraded by the addition of 1 μL RQ1 DNase and digestion at 37 °C for 15 minutes. Transcribed crRNAs were subsequently purified with the miRNeasy Mini Kit (Qiagen). In vitro cleavage reactions consisted of 2.5 nM PvuI-linearized substrate plasmid, 300 nM crRNA, and 200 nM purified Cas12a protein in cleavage buffer (10 mM Hepes pH 7.5, 150 mM NaCl and 5 mM MgCl2), and were performed at 37 °C unless otherwise indicated. Plasmid substrates for temperature tolerance assays encoded the PAMDA site 2 spacer with a TTTA PAM (in Fig. 2b and Supplementary Fig. 7k). Cleavage reaction master-mixes were prepared and then aliquoted into 5 μL volumes for each time point, incubated in a thermal cycler, and halted by the addition of 10 μL of stop buffer (0.5% SDS and 50 mM EDTA). Stopped aliquots were purified with paramagnetic beads and the percent cleavage was quantified by QIAxcel ScreenGel Software (v1.4 or v 1.5).
PAM determination assay.
Plasmid libraries encoding target sites with randomized sequences were cloned using Klenow(-exo) (NEB) to fill in the bottom strands of two separate oligos harboring 10 nt randomized sequences 5’ of two distinct spacer sequences (Supplementary Table 7). The double-stranded product was digested with EcoRI and ligated into EcoRI and SphI digested p11-lacY-wtx1 (Addgene plasmid 69056; a gift from Huimin Zhao). Ligations were transformed into electrocompetent XL1 Blue E. coli, recovered in 9 mL of SOC at 37 °C for 1 hour, and then grown for 16 hours in 150 mL of LB medium with 100 mg/L carbenicillin. The complexity of each library was estimated to be greater than 106 based on the number of transformants. Prior to use in in vitro cleavage reactions, plasmid libraries were linearized with PvuI (NEB).
Cleavage reactions of the randomized PAM plasmid libraries were performed as described above, with aliquots stopped at 3, 6, 12, 24, and 48 minutes. Reactions were purified with magnetic beads and approximately 1-5 ng of purified plasmid was used as template for PCR amplification of uncleaved molecules with Phusion DNA Polymerase (NEB) for 15 cycles. PCR primers encode a 4 nt barcode upstream of the PAM to enable demultiplexing of the time-point samples. Amplicons were also generated from the untreated plasmids to determine initial PAM representation in the libraries. Purified PCR products were quantified with QuantiFluor dsDNA System (Promega), normalized, and pooled for library preparation with Illumina dual-indexed adapters using a KAPA HTP PCR-free Library Preparation Kit (KAPA BioSystems). Libraries were quantified using the Universal KAPA Illumina Library qPCR Quantification Kit (KAPA Biosystems) and sequenced on an Illumina MiSeq sequencer using a 300-cycle v2 kit (Illumina).
Sequencing reads were analyzed using a custom Python script (available upon request) to estimate cleavage rates on each PAM for a given protein (Supplementary Table 1). Paired-end reads were filtered by Phred score (≥Q30) and then merged with the requirement of perfect matches of time point barcodes, PAM, and spacer sequence. Counts were generated for every 4 and 5 nt PAM for all time points, protein, and spacer. PAM counts were then corrected for inter-sample differences in sequencing depth, converted to a fraction of the initial representation of that PAM in the original plasmid library (as determined by the untreated control), and then normalized to account for the increased fractional representation of uncut substrates over time due to depletion of cleaved substrates (by selecting the 5 PAMs with the highest average counts across all time points to represent the profile of uncleavable substrates). The depletion of each PAM over time was then fit to an exponential decay model (y(t) = Ae−kt, where y(t) is the normalized PAM count, t is the time (minutes), k is the rate constant, and A is a constant), by linear least squares regression.
Targeting range calculations.
The targeting ranges of wild-type and variant AsCas12a nucleases were assessed on various annotated genomic elements using GENCODE’s Release 27 GTF file. Complete occurrences of targetable 4 nt PAMs were enumerated within regions encompassing 1 kb upstream of all transcription start sites (TSSs), within the first exon of all genes, and within all annotated miRNAs. Parameter value(s) for each element in the GTF file were: Exon1, feature-type exon, exon_number 1, gene_type protein_coding; TSS, feature-type transcript, gene_type protein_coding or miRNA; miRNA, feature-type gene, gene_type miRNA. For each element, PAM counts were normalized by length and were visualized through a boxplot. The PAM identification and enumeration script will be made available upon request. Targetable PAMs for Cas12a nucleases included: TTTV, for wild-type AsCas12a; TTYN, RTTC, CTTV, TATM, CTCC, TCCC, TACA (tier 1), and RTTS, TATA, TGTV, ANCC, CVCC, TGCC, GTCC, TTAC (tier 2) PAMs for enAsCas12a (see Fig. 1e and Supplementary Fig. 3h); TATV, AsCas12a-RVR; and TYCV for AsCas12a-RR3.
Statistics
Statistical significance between data sets was calculated using Wilcoxon signed-rank or Mann-Whitney tests using GraphPad Prism version 7.0c (see results of tests in Supplementary Table 8). A multiple comparisons adjustment was performed using the Bonferroni correction (34 total tests; P < 0.00147). P values are reported using GraphPad style: not significant (ns), P > 0.05; *, P < 0.05; **, P < 0.01; ***, P <0.001; ****, P <0.0001.
Code Availability
The custom Python script for PAMDA data analysis, and the PAM identification and enumeration script, will be made available upon request.
Data Availability and Accession Code Availability Statements
Data sets from GUIDE-seq and high-throughput sequencing experiments (for PAMDA and base editing experiments) have been deposited with the National Center for Biotechnology Information Sequence Read Archive under BioProject ID PRJNA508751.
Supplementary Material
Acknowledgements
We thank J. Grünewald for preparing paramagnetic beads, S. Garcia for informatics support, and members of the Joung and Maus laboratories for advice. This work was supported by the Desmond and Ann Heathwood MGH Research Scholar Award (to J.K.J.), Natural Sciences and Engineering Research Council of Canada (NSERC) Banting and Charles A. King Trust Postdoctoral Fellowships (B.P.K.), the National Institutes of Health (NIH) Awards K99 CA218870 (B.P.K.), R00 HG008399 (L.P.), R35 GM118158 (M.J.A. and J.K.J.), and RM1 HG009490 (J.K.J.), the Bill & Melinda Gates Foundation OPP1159968 (J.K.J.), and an award from the Massachusetts General Hospital Collaborative Center for X-Linked Dystonia-Parkinsonism. Plasmids described in this work are available through the non-profit plasmid repository Addgene (http://www.addgene.org/crispr-cas).
Footnotes
Competing Financial Interests Statement
B.P.K is a scientific advisor to Avectas. J.K.J. is a member of the Board of Directors of the American Society of Gene and Cell Therapy. J.K.J. has financial interests in Beam Therapeutics, Blink Therapeutics, Editas Medicine, Endcadia, Monitor Biotechnologies (formerly known as Beacon Genomics), Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. M.J.A. has financial interests in Monitor Biotechnologies. J.K.J. holds equity in EpiLogic Therapeutics. J.K.J.’s and M.J.A.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. B.P.K., A.A.S., and J.K.J. are inventors on patent applications covering the Cas12a variants described in this work. Y.E.T., B.P.K., and J.K.J. are inventors on a patent application describing Cas12-based transcriptional activators. M.V.M. reports personal fees from Adaptimmune, personal fees from Adaptive Biotechnologies, grants and personal fees from Agentus, personal fees from Bluebird Bio, personal fees from Cellectis, grants and personal fees from CRISPR Therapeutics, personal fees from Incysus, grants and personal fees from Kite Pharma, personal fees from Juno, personal fees from MPM, personal fees from Novartis, personal fees from Takeda, grants and personal fees from TCR2 (SAB), personal fees from Third Rock Ventures, personal fees from Windmil (SAB), personal fees from Century, outside the submitted work. M.V.M. has a patent related to CAR T cells for multiple myeloma, lymphoma, and glioblastoma (none of which are the subject of this manuscript) pending.
References:
- 1.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim HK et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods 14, 153–159 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Gao L et al. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol 35, 789–792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu PD, Lander ES & Zhang F Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez AA, Lim WA & Qi LS Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol 17, 5–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright AV, Nuñez JK & Doudna JA Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 164, 29–44 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Komor AC, Badran AH & Liu DR CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 168, 20–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov S et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shmakov S et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol 15, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinstiver BP et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol 34, 869–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonfara I, Richter H, Bratovič M, Le Rhun A & Charpentier E The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Zetsche B et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol 35, 31–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tak YE et al. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods 14, 1163–1166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong G, Wang H, Li Y, Tran MH & Farzan M Cpf1 proteins excise CRISPR RNAs from mRNA transcripts in mammalian cells. Nat. Chem. Biol 13, 839–841 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow RD, Wang G, Codina A, Ye L & Chen S Mapping in vivo genetic interactomics through Cpf1 crRNA array screening. (2017). doi: 10.1101/153486 [DOI] [Google Scholar]
- 16.Kim D et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol 34, 863–868 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Yamano T et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 165, 949–962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao P, Yang H, Rajashankar KR, Huang Z & Patel DJ Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 26, 901–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karvelis T et al. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 16, 253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno-Mateos MA et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun 8, 2024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3, 17018 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Kim H et al. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8, 14406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y et al. Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nat Commun 8, 2095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X et al. Gene activation in human cells using CRISPR/Cpf1-p300 and CRISPR/Cpf1-SunTag systems. Protein Cell 9, 380–383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X et al. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov 3, 17018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavez A et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol 36, 324–327 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729–aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Komor AC, Badran AH & Liu DR Editing the Genome Without Double-Stranded DNA Breaks. ACS Chem. Biol 13, 383–388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan WX et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8, 15058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature (2016). doi: 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JS et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. (2017). doi: 10.1101/160036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, Kim D, Cho SW, Kim J & Kim J-S Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang X et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol 208, 44–53 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Zuris JA et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol 33, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohland N & Reich D Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyon D et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol 30, 460–465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vouillot L, Thélie A & Pollet N Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5, 407–415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sentmanat MF, Peters ST, Florian CP, Connelly JP & Pruett-Miller SM A Survey of Validation Strategies for CRISPR-Cas9 Editing. Sci Rep 8, 888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HK et al. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol 36, 239–241 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Clement K et al. Analysis and comparison of genome editing using CRISPResso 2 (2018). doi: 10.1101/392217 [DOI] [Google Scholar]
- 45.Kleinstiver BP et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai SQ, Topkar VV, Joung JK & Aryee MJ Open-source guideseq software for analysis of GUIDE-seq data. Nat. Biotechnol. 34, 483–483 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Studier FW Stable expression clones and auto-induction for protein production in E. coli. Methods Mol. Biol 1091, 17–32 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Maeder ML et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishimasu H et al. Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1. Mol. Cell 67, 139–147.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets from GUIDE-seq and high-throughput sequencing experiments (for PAMDA and base editing experiments) have been deposited with the National Center for Biotechnology Information Sequence Read Archive under BioProject ID PRJNA508751.