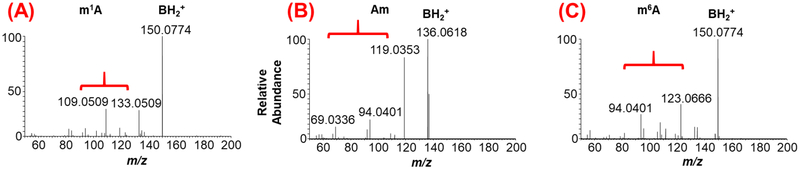
Figure 4:
Differentiation of positional isomers of methylated adenosines (m/z 282.12) through molecular fingerprints generated by higher-energy collisional dissociation (HCD) analysis. (A) HCD of m1A indicating the presence of modified nitrogenous base and its fragments. (B) HCD of Am depicting the unmodified adenine and its fragment ions following the loss of methylated ribose. (C) HCD of m6A depicting the modified nitrogenous base and its fragment ions. The fragment ions are indicated by curved parenthesis. Note the molecular fingerprint of nitrogenous base fragment ions is different for each positional isomer.