FIGURE 2. MEK2 regulates IL-1β and HIF-1α expression at the transcript and protein level.
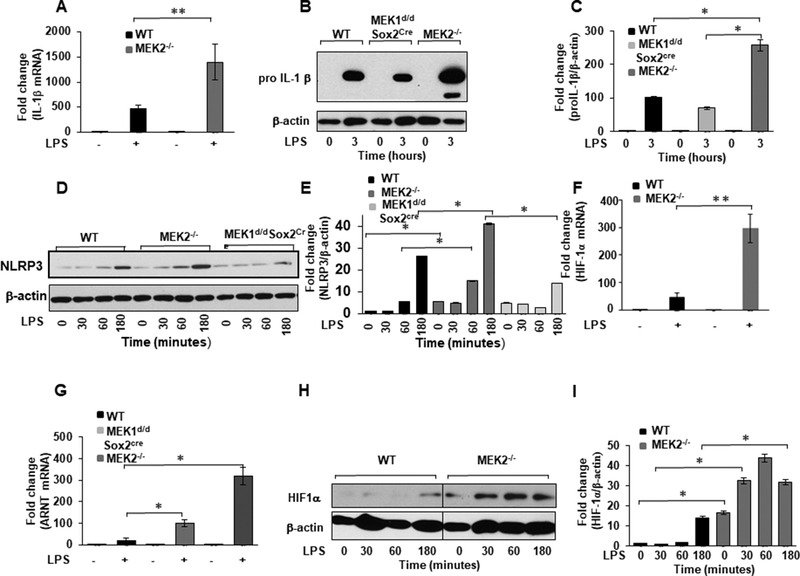
BMDMs derived from WT and MEK2−/− and Mek1d/d Sox2Cre mice were treated with LPS (100 ng/mL) for 1h. Total RNA was extracted and reverse-transcribed using the Reverse Transcription System. (A) The primers targeting IL-1β were used to amplify cDNA using iQSYBR Green Supermix. Relative mRNA levels were calculated by normalizing to GAPDH. Data were analyzed using the paired, two-tailed Student’s t test, and the results were expressed as fold change. (B) BMDMs derived from WT and MEK2−/− and Mek1d/d Sox2Cre mice were treated with LPS (100 ng/mL) for 3h. Whole cell extracts were prepared and subjected to SDS-PAGE and Western blot analysis using specific antibodies against pro IL-1β. (C) Equal loading was determined using antibodies against β-actin. (D) BMDMs derived from WT and MEK2−/− and Mek1d/d Sox2Cre were treated with LPS (100 ng/mL) for different time points as indicated. Whole cell extracts were prepared and subjected to SDS-PAGE and Western analysis using specific antibodies to NLRP3. (E) Densitometric values expressed as fold changes of the ratio NLRP3/ β-actin. (F) HIF-1α mRNA expression. RNA was isolated from cells and expression assessed using qRT-PCR. Values were normalized to GAPDH. Results represent mean values of 4 independent experiments. Using ANOVA Mann-Whitney U test a p value of <0.05 was considered significant and error bars indicate SEM. (G) ARNT (HIF-1 β) mRNA expression. Total RNA was extracted from WT and MEK2−/− and Mek1d/d Sox2Cre BMDMs treated with LPS (100 ng/mL) for 1h. The primers targeting ARNT were used to amplify cDNA. Relative mRNA levels were calculated by normalizing to GAPDH. Data were analyzed using the paired, two-tailed Student’s t test, and the results were expressed as fold change. (H) HIF-1α expression. Whole cell extracts were subjected to SDS-PAGE and Western blot analysis using specific antibodies against HIF-1α. Equal loading was determined using antibodies against β-actin. (I) Densitometric analysis of at least 3 independent experiments expressed as fold change of the ratio HIF-1α/ β-actin.
