Abstract
Objective
Accumulating reports reveal that serving as an oncogenic factor LAMTOR5 is involved in the progression of many specific cancers. Glucose transporter 1 (GLUT1) is frequently identified in many cancers. However, it remains unexplored whether GLUT1 plays a role in LAMTOR5-enhanced liver cancer. Here, we aim to decipher the function of LAMTOR5 in the regulation of GLUT1 in liver cancer.
Methods
The effect of LAMTOR5 on GLUT1 was analyzed using Western blotting and RT-PCR assay. Dose-increased over-expression or silencing of LAMTOR5 was performed through transient transfection. LAMTOR5-activated GLUT1 promoter was revealed by luciferase reporter assay. The regulation of GLUT1 by LAMTOR5/NF-κB was examined via Western blotting and luciferase reporter assays.
Results
The data showed that in liver cancer cells under the administration with dose-increased LAMTOR5, the level of mRNA and protein of GLUT1 was obviously raised. Our data revealed that the activities of GLUT1 promoter were induced by LAMTOR5. Then, we found that the elevation of GLUT 1 mediated by LAMTOR5 slowed when the inhibitor or siRNAs of NF-κB was introduced into the liver cancer cells. Conclusion. LAMTOR5 is responsible for the activation of GLUT1 via transcription factor NF-κB in liver cancer.
Keywords: LAMTOR5, GLUT1, NF-κB, liver cancer
1. Introduction
Liver cancer is one of major causes of cancer-associated death throughout the world [1, 2, 3]. The incidence of liver cancer has been increasing in European and American countries [4]. LAMTOR5 (also known as hepatitis B X-interacting protein, HBXIP) is firstly identified because of its interaction with HBX protein [5], and its constitutive expression is revealed in a great number of tissues. A report reveals that as one memberof a regulator complex consisting of p18, MP1, p14, LAMTOR5 and C7orf59, LAMTOR5 plays a significant role in amino acids-induced mTORC1 activation [6]. During the development of various cancers including lung cancer, breast cancer, gastric cancer, bladder cancer, ovarian cancer, or liver cancer, LAMTOR5 can function as an oncogenic factor [7, 8, 9, 10, 11, 12, 13, 14, 15, 16]. Over-expressed LAMTOR5 is capable of enhancing the proliferation, migration or abnormal glucose metabolism of cancer cells [17, 18, 19]. Yet, investigation is still required for the detailed mechanism by which LAMTOR5 is involved in cancer progression.
Abnormal glucose metabolism, such as Warburg effect, is one crucial part of the hallmarks of cancer [20]. It has been revealed that the elevated aerobic glycolysis is closely associated with the progression of liver cancer [21]. Aerobic metabolism includes many features, such as ROS [22]. Glucose transporter 1 (GLUT1) functions in the glucose transport across the cellular plasma membranes [23]. Compared with normal tissues, elevated GLUT1 is frequently found in great numbers of caners [24, 25]. It has been reported that GLUT1 augmentation is positively correlated with the malignant progression of liver cancer [26, 27]. It is still unclear whether LAMTOR5 affects liver cancer progression through modulating the expression of GLUT1.
In our present study, we aim to decipher the function and mechanism involved in LAMTOR5-induced GLUT1 in liver cancer. Notably, we disc lose that LAMTOR5 is able to upregulate the expression of GLUT1 in liver cancer cells, in which NF-κB as a famous transcription factor is responsible for the activation of GLUT1 induced by LAMTOR5. Our findings could potentially provide a more detailed mechanism of LAMTOR5-regulated GLUT1 and more therapeutic targets for liver cancer.
2. Materials and methods
2.1. Cell culture
Followed by the protocol of ATCC, human hepatoma cell line, HepG2 grew in DMEM medium with 10% fetal bovine serum.
2.2. Plasmids and reagents
According to the report30 we cloned the promoter region of GLUT1 into the KpnI/HindⅢ site within the pGL3-basic vector (Promega, USA). pGL3-Basic activities were normalized by pRL-TK. RiboBio (Guangzhou, China) is responsible for the synthesis of siRNAs targeting LAMTOR5 or NF-κB.
2.3. Reverse transcription-polymerase chain reaction (RT-PCR)
From liver cancer cells total RNA was acquired by using TRIzol Reagent (Invitrogen, USA). ImPro-II Reverse Transcriptase (Promega, USA) was applied in reverse transcription reaction. GAPDH was used as loading control.
2.4. Western blotting
Liver cancer HepG2 cells were lysed by RIPA buffer and the total protein was then extracted. Post electrophoresis total protein was transferred from SDS-PAGE gel to PVDF membranes (ThermoFisher Scientific, USA). Anti-GLUT1 (Abcam, USA) or anti-β-actin (Abcam, USA) was used as the primary antibodies in this study.
2.5. Luciferase reporter analysis
HepG2 cells were plated into 24-well plates. The cells were co-transfected with reporter gene plasmids and pRL-TK plasmid (Promega, USA) and corresponding vectors, siRNAs, or inhibitors. The cells were collected after 48 hours and the luciferase activity was quantified according to the manufacturer’s instructions provided by Promega. Each experiment was repeated at least three times.
2.6. Patient samples
Twenty-one HCC tissues were provided by the General Hospital of Tianjin Medical University (Tianjin, China) from HCC patients by the surgical department (Supplementary Table S1). Written consents approving the tissue use in study were gained from patients. Study protocol has been approved by the Research Ethics Board at the General Hospital of Tianjin Medical University (Tianjin, China).
2.7. Statistical analysis
The results were expressed as means ± standard error of the mean (SEM). Statistical differences between the two groups were analyzed by Student’s t-test. Non-significant difference was marked with ns. Criterions for statistically significant differences were considered as followed: **P<0.01; ***P<0.001. The correlation between GLUT1 and LAMTOR5 in human HCC samples was evaluated by Pearson’s correlation coefficient.
3. Results
3.1. Oncogenic LAMTOR5 is able to upregulate the expression of GLUT1 in hepatoma cells
Accumulating evidence has revealed that some tumor-associated proteins are involved in LAMTOR5-promoted cancers [28, 29]. As a glucose transporter, GLUT1 is overexpressed in numerous cancers and closely associated with the development of many cancers. Here, we are interested in the role of GLUT1 in LAMTOR5-related liver cancer. When LAMTOR5 was introduced into liver cancer HepG2 cells, we tested the alteration of GLUT1 expression by using reverse transcription-polymerase chain reaction (RT-PCR) and Western blotting. Our data demonstrated that the level of mRNA and protein of GLUT1 was increased in liver cancer cells under different doses of LAMTOR5 transfection (Fig. 1A, B). Thus, we indicate that in liver cancer, oncogenic LAMTOR5 can induce GLUT1 expression.
Figure 1.
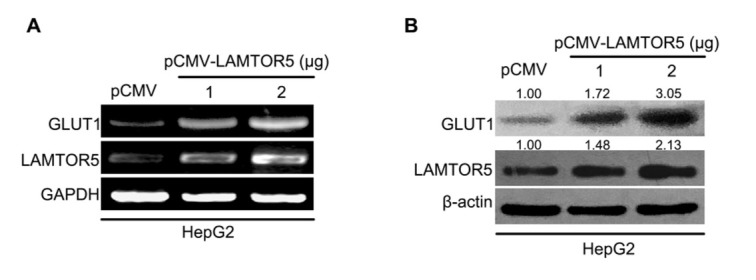
Oncogenic LAMTOR5 is able to upregulate the expression of GLUT1 in hepatoma cells. (A, B) Level of mRNA and protein of GLUT1 in hepatoma HepG2 cells after LAMTOR5 was overexpressed was analyzed through RT-PCR and Western blotting. ImageQuant 5.2 software (GE Healthcare, UK) was used to quantify the bands of GLUT1 and LAMTOR5 relative to β-actin.
3.2. LAMTOR5 is capable of activating the transcription of GLUT1
To clarify how LAMTOR5 upregulates GLUT1, we cloned the full-length promoter of GLUT1 (-1306/+197) according to previous reports [30] and determined its activities in liver cancer cells. After over-expression of LAMTOR5 was performed, the activities of the GLUT1 promoter in HepG2 cells were analyzed by luciferase reporter assay. We observed that elevated dose of LAMTOR5 could enhance the activities of GLUT1 promoter (Fig. 2A). At the same time, RNA interference (RNAi) targeting GLUT1 mRNA was able to dose-dependently inhibit the activities of GLUT1 promoter (Fig. 2B). So, our data shows that GLUT1 promoter can be stimulated by oncogenic LAMTOR5 in liver cancer cells.
Figure 2.
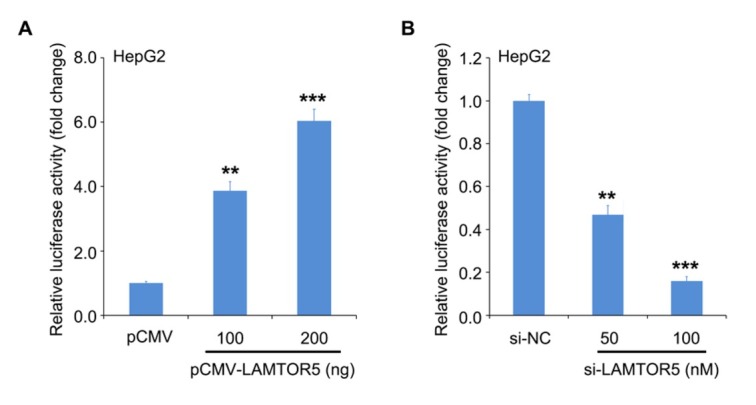
LAMTOR5 is capable of activating the transcription of GLUT1. (A, B) The effect of LAMTOR5 on GLUT1 promoter was determined using luciferase reporter assay in HepG2 cells transiently transfected with LAMTOR5 vector or siRNAs. **P < 0.01, ***P < 0.001.
3.3. LAMTOR5 upregulates NF-κB to promote the transcription of GLUT1 in liver cancer
Based on the findings that binding sites of transcription factor might exist, NF-κB within the promoter region of GLUT1 and LAMTOR5 is able to upregulate NF-κB in cancer cells [9, 30, 31], we next tried to investigate whether NF-κB is responsible for LAMTOR5-induced GLUT1 transcription in the cells. We found that LAMTOR5 could obviously promote the activities of the GLUT1 promoter, but it lost the effect on the mutant GLUT1 promoter in which the binding sites of NF-κB was mutated. At the same time, LAMTOR5-increased activities of GLUT1 promoter could be retarded under the treatment with the inhibitor of NF-κB, PDTC (Fig. 3A). Furthermore, we synthesized small interference RNA (siRNA) targeting NF-κB and transfected it into liver cancer cells. The data revealed that over-expressed LAMTOR5 was capable of inducing the expression of NF-κB and thereafter its downstream target gene, GLUT1 using an immunoblotting assay. After LAMTOR5 was knockdown by siRNAs, the augmentation of NF-κB and GLUT1 ceased in the liver cancer cells (Fig. 3B). Altogether, our data imply that NF-κB is required for LAM-TOR5-activated GLUT1 in liver cancer.
Figure 3.
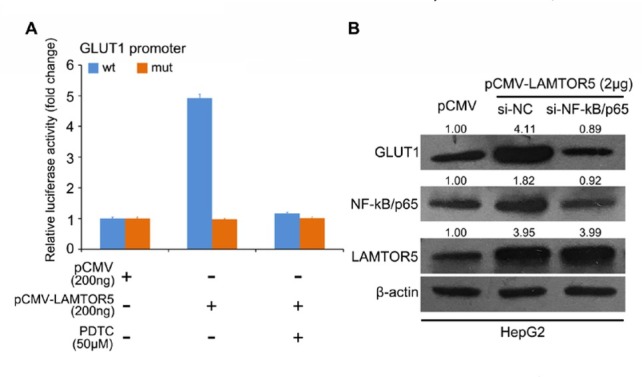
LAMTOR5 upregulates NF-κB to promote the transcription of GLUT1 in liver cancer. (A) The effect of LAMTOR5 and/or NF-κB inhibitor (PDTC) on wild type (wt) and mutant (mut) GLUT1 promoter was evaluated. (B) The level of NF-κB and GLUT1 in liver cancer cells under the administration of LAMTOR5 overexpression and/or NF-κB/p65 siRNAs was evaluated through Western blotting. ImageQuant 5.2 software (GE Healthcare, UK) was used to quantify the bands of GLUT1, NF-κB or LAMTOR5 relative to β-actin. ***P < 0.001.
3.4. Overexpressed GLUT1 is associated with high level of LAMTOR5 in clinical HCC tissues
Finally, we used twenty-one cases of clinical HCC tissues and paired noncancerous tissues to evaluate the correlation between GLUT1 and LAMTOR5. We observed that GLUT1 was positively correlated with LAMTOR5 in clinical HCC samples (R2=0.8311, p < 0.01) (Fig. 4). Our data imply that GLUT1 is positively correlated to with LAMTOR5 in clinical HCC tissues.
Figure 4.
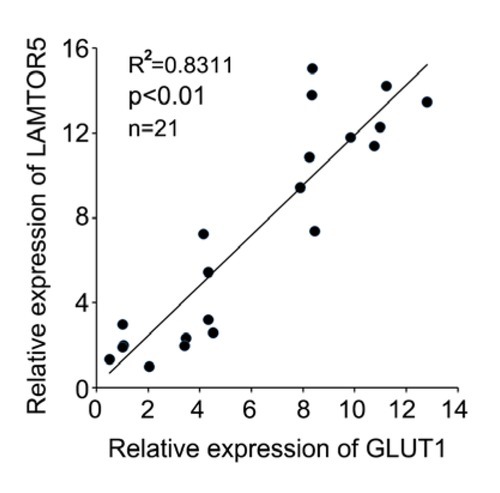
Overexpressed GLUT1 is associated with high level of LAMTOR5 in clinical HCC tissues. The correlation between GLUT1 and LAMTOR5 was evaluated in twenty-one cases of clinical HCC samples (Pearson′s correlation coefficient, R2= 0.8311, **P < 0.01).
4. Discussion
Liver cancer remains one of main causes of tumor-associated death around the world [1, 2, 3]. Among the European and American countries, the incidence of liver cancer has been increasing [4]. LAMTOR5 is firstly identified because of its interaction with HBX protein [5], and its constitutive expression is revealed in a great number of tissues. As one member of a regulator complex consisting of p18, MP1, p14, LAMTOR5 and C7orf59, LAMTOR5 can take great part in amino acids-induced mTORC1 activation [6]. During the development of many cancers, LAMTOR5 can function as an oncogenic factor [7, 8, 9, 10, 11, 12, 13, 14, 15, 16]. Over-expressed LAMTOR5 can affect cell proliferation, migration or abnormal glucose metabolism in cancers [17, 18, 19]. Here, we are interested in the role of GLUT1 in LAMTOR5-mediated liver cancer.
Abnormal glucose metabolism, such as Warburg effect, is one hallmark of cancer [20]. The elevated aerobic glycolysis is closely associated with the progression of liver cancer [21]. GLUT1 can transport the glucose across the cellular plasma membranes [23]. Highly expressed GLUT1 is frequently revealed in many types of cancers [24, 25]. Its augmentation is closely related to the malignant progression of liver cancer [26, 27].
To decipher the role of GLUT1 in LAMTOR5-related liver cancer, we transfected the vector containing full-length LAMTOR5 into liver cancer HepG2 cells. Then, we tested the level of GLUT1 in the cells using Western blotting and RT-PCR analysis. Our results manifested that not only the mRNA level, but also the protein level for GLUT1 was enhanced by oncogenic LAMTOR5 in liver cancer cells. It indicates that LAMTOR5 involves the regulation of GLUT1 in liver cancer. To further determine whether LAMTOR5 could regulate the promoter activity of GLUT1, we cloned this promoter region into pGL3-basic vector. Our data showed that the luciferase activity critically increased along with the elevated dose of LAMTOR5 in HepG2 cell line. Furthermore, the luciferase activity obviously declined when LAMTOR5 was silenced by siRNAs. In the further investigation, we focus on the dissecting the mechanism by which LAMTOR5 activates the transcription of GLUT1 in liver cancer. Previous reports have found that binding sites of transcription factor, NF-κB , may exist within the promoter region of GLUT1. Thereby, LAMTOR5 is able to upregulate NF-κB in cancer cells [9, 30, 31]. Thus, we next cloned the mutant GLUT1 promoter containing mutated binding site of NF-κB. We found that LAMTOR5 inability to induce GLUT1 activation when the binding site of NF-κB within GLUT1 promoter region was mutated. An inhibitor of NF-κB, PDTC was able to abrogate enhancement of the promoter activities of GLUT1 by LAMTOR5. Meanwhile, after the administration of NF-κB siRNAs the promotion of GLUT1 by LAMTOR5 was retarded in liver cancer cells. We finally confirmed the positive correlation between GLUT1 with LAMTOR5 in clinical liver cancer tissues.
5. Conclusion
For our current study, we uncovered that oncogenic LAMTOR5 is able to induce the expression of GLUT1 through increasing transcription factor NF-κB during the development of liver cancer. Our study will shed light on the function of GLUT1 in LAMTOR5-related liver cancer.
Footnotes
Funding: This work was supported by The Science & Technology Development Fund of the Tianjin Education Commission for Higher Education (No.2017KJ196).
Conflict of interest
Conflicts of interest: The authors have no conflicts of interest to declare.
Supplementary Table 1.
Clinical characteristics of liver cancer samples
| No. | Sex | Age | Pathology diagnosis |
|---|---|---|---|
| 1 | M | 42 | liver cancer |
| 2 | M | 61 | liver cancer |
| 3 | M | 52 | liver cancer |
| 4 | M | 50 | liver cancer |
| 5 | M | 47 | liver cancer |
| 6 | M | 54 | liver cancer |
| 7 | M | 56 | liver cancer |
| 8 | M | 49 | liver cancer |
| 9 | M | 63 | liver cancer |
| 10 | M | 59 | liver cancer |
| 11 | M | 70 | liver cancer |
| 12 | M | 55 | liver cancer |
| 13 | M | 46 | liver cancer |
| 14 | M | 67 | liver cancer |
| 15 | M | 52 | liver cancer |
| 16 | M | 65 | liver cancer |
| 17 | M | 46 | liver cancer |
| 18 | M | 52 | liver cancer |
| 19 | M | 59 | liver cancer |
| 20 | M | 64 | liver cancer |
| 21 | M | 51 | liver cancer |
References
- [1].El-Serag H.B., Rudolph K.L.. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557. doi: 10.1053/j.gastro.2007.04.061. –. [DOI] [PubMed] [Google Scholar]
- [2].Forner A, Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245. doi: 10.1016/S0140-6736(11)61347-0. –. [DOI] [PubMed] [Google Scholar]
- [3].Ma C, Kesarwala A.H., Eggert T, Medina-Echeverz J, Kleiner D.E., Jin P. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253. doi: 10.1038/nature16969. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Endig J, Buitrago-Molina L.E., Marhenke S, Reisinger F, Saborowski A, Schutt J. Dual Role of the Adaptive Immune System in Liver Injury and Hepatocellular Carcinoma Development. Cancer cell. 2016;30:308. doi: 10.1016/j.ccell.2016.06.009. et al. –. [DOI] [PubMed] [Google Scholar]
- [5].Melegari M, Scaglioni P.P., Wands J.R. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J Virol. 1998;72:1737. doi: 10.1128/jvi.72.3.1737-1743.1998. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bar-Peled L, Schweitzer L.D., Zoncu R, Sabatini D.M.. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196. doi: 10.1016/j.cell.2012.07.032. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fujii R, Zhu C, Wen Y, Marusawa H, Bailly-Maitre B, Matsuzawa S. HBXIP, cellular target of hepatitis B virus oncoprotein, is a regulator of centrosome dynamics and cytokinesis. Cancer Res. 2006;66:9099. doi: 10.1158/0008-5472.CAN-06-1886. et al. –. [DOI] [PubMed] [Google Scholar]
- [8].Krepela E, Dankova P, Moravcikova E, Krepelova A, Prochazka J, Cermak J. Increased expression of inhibitor of apoptosis proteins, survivin and XIAP, in non-small cell lung carcinoma. Int J Oncol. 2009;35:1449. doi: 10.3892/ijo_00000464. et al. –. [DOI] [PubMed] [Google Scholar]
- [9].Hu N, Zhang J, Cui W, Kong G, Zhang S, Yue L. miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J Biol Chem. 2011;286:13714. doi: 10.1074/jbc.M110.204131. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang F, Fei H, Qi B, Yao S, Chang Z. Overexpression of hepatitis B x-interacting protein in HepG2 cells enhances tumor-induced angiogenesis. Mol Cell Biochem. 2012;364:165. doi: 10.1007/s11010-011-1215-5. –. [DOI] [PubMed] [Google Scholar]
- [11].Zhang H, Xu F, Xie T, Jin H, Shi L. β-elemene induces glioma cell apoptosis by downregulating survivin and its interaction with hepatitis B X-interacting protein. Oncol Rep. 2012;28:2083. doi: 10.3892/or.2012.2022. –. [DOI] [PubMed] [Google Scholar]
- [12].Liu F, You X, Wang Y, Liu Q, Liu Y, Zhang S. The oncoprotein HBXIP enhances angiogenesis and growth of breast cancer through modulating FGF8 and VEGF. Carcinogenesis. 2014;35:1144. doi: 10.1093/carcin/bgu021. et al. –. [DOI] [PubMed] [Google Scholar]
- [13].Xu F, Zhu X, Han T, You X, Liu F, Ye L. The oncoprotein hepatitis B X-interacting protein promotes the migration of ovarian cancer cells through the upregulation of S-phase kinase-associated protein 2 by Sp1. Int J Oncol. 2014;45:255. doi: 10.3892/ijo.2014.2411. et al. –. [DOI] [PubMed] [Google Scholar]
- [14].Cheng D, Liang B, Li Y. HBXIP expression predicts patient prognosis in breast cancer. Med Oncol. 2014;31:210. doi: 10.1007/s12032-014-0210-6. [DOI] [PubMed] [Google Scholar]
- [15].Li X, Liu S. Suppression of HBXIP Reduces Cell Proliferation, Migration and Invasion In Vitro, and Tumorigenesis In Vivo in Human Urothelial Carcinoma of the Bladder. Cancer Biother Radiopharm. 2016;31:311. doi: 10.1089/cbr.2016.2038. –. [DOI] [PubMed] [Google Scholar]
- [16].Piao J.J., Li N, Wang Y.X., Lin Z.H., Liu S.P. HBXIP expression in gastric adenocarcinoma predicts poor prognosis. Zhonghua bing li xue za zhi = Chinese journal of pathology. 2017;46:88. doi: 10.3760/cma.j.issn.0529-5807.2017.02.005. –. [DOI] [PubMed] [Google Scholar]
- [17].Liu S, Li L, Zhang Y, Zhang Y, Zhao Y, You X. The oncoprotein HBXIP uses two pathways to up-regulate S100A4 in promotion of growth and migration of breast cancer cells. J Biol Chem. 2012;287:30228. doi: 10.1074/jbc.M112.343947. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shi H, Li Y, Feng G, Li L, Fang R, Wang Z. The oncoprotein HBXIP up-regulates FGF4 through activating transcriptional factor Sp1 to promote the migration of breast cancer cells. Biochem Biophys Res Commun. 2016;471:89. doi: 10.1016/j.bbrc.2016.01.174. et al. –. [DOI] [PubMed] [Google Scholar]
- [19].Liu F, Zhang W, You X, Liu Y, Li Y, Wang Z. The oncoprotein HBXIP promotes glucose metabolism reprogramming via downregulating SCO2 and PDHA1 in breast cancer. Oncotarget. 2015;6:27199. doi: 10.18632/oncotarget.4508. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu J, Zhang C, Wu R, Lin M, Liang Y, Liu J. RRAD inhibits the Warburg effect through negative regulation of the NF-kappaB signaling. Oncotarget. 2015;6:14982. doi: 10.18632/oncotarget.3719. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martinez-Outschoorn UE, Peiris-Pages M, Pestell R.G., Sotgia F, Lisanti M.P.. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:113. doi: 10.1038/nrclinonc.2017.1. [DOI] [PubMed] [Google Scholar]
- [22].Poyton R.O., Ball K.A., Castello P.R. Mitochondrial generation of free radicals and hypoxic signaling. Trends Endocrinol Metab. 2009;20:332. doi: 10.1016/j.tem.2009.04.001. –. [DOI] [PubMed] [Google Scholar]
- [23].Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34:121. doi: 10.1016/j.mam.2012.07.001. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Airley R.E., Mobasheri A. Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: novel pathways and targets for anticancer therapeutics. Chemotherapy. 2007;53:233. doi: 10.1159/000104457. –. [DOI] [PubMed] [Google Scholar]
- [25].Medina R.A., Owen G.I. Glucose transporters: expression, regulation and cancer. Biol Res. 2002;35:9. doi: 10.4067/s0716-97602002000100004. –. [DOI] [PubMed] [Google Scholar]
- [26].Rothwell P.M., Wilson M, Price J.F., Belch J.F., Meade T.W., Mehta Z. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet. 2012;379:1591. doi: 10.1016/S0140-6736(12)60209-8. –. [DOI] [PubMed] [Google Scholar]
- [27].Huang Q, Li J, Xing J, Li W, Li H, Ke X. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J Hepatol. 2014;61:859. doi: 10.1016/j.jhep.2014.04.035. et al. –. [DOI] [PubMed] [Google Scholar]
- [28].Zhao Y, Li H, Zhang Y, Li L, Fang R, Li Y. Oncoprotein HBXIP Modulates Abnormal Lipid Metabolism and Growth of Breast Cancer Cells by Activating the LXRs/SREBP-1c/FAS Signaling Cascade. Cancer Res. 2016;76:4696. doi: 10.1158/0008-5472.CAN-15-1734. et al. –. [DOI] [PubMed] [Google Scholar]
- [29].Xu F, You X, Liu F, Shen X, Yao Y, Ye L. The oncoprotein HBXIP up-regulates Skp2 via activating transcription factor E2F1 to promote proliferation of breast cancer cells. Cancer Lett. 2013;333:124. doi: 10.1016/j.canlet.2013.01.029. et al. –. [DOI] [PubMed] [Google Scholar]
- [30].Liu Y.X., Feng J.Y., Sun M.M., Liu B.W., Yang G, Bu Y.N.. Aspirin inhibits the proliferation of hepatoma cells through controlling GLUT1-mediated glucose metabolism. Acta Pharmacol Sin. 2019;40:122. doi: 10.1038/s41401-018-0014-x. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang F.Z., Sha L, Zhang W.Y., Wu L.Y., Qiao L, Li N. Involvement of hepatitis B X-interacting protein (HBXIP) in proliferation regulation of cells. Acta Pharmacol Sin. 2007;28:431. doi: 10.1111/j.1745-7254.2007.00531.x. et al. –. [DOI] [PubMed] [Google Scholar]


