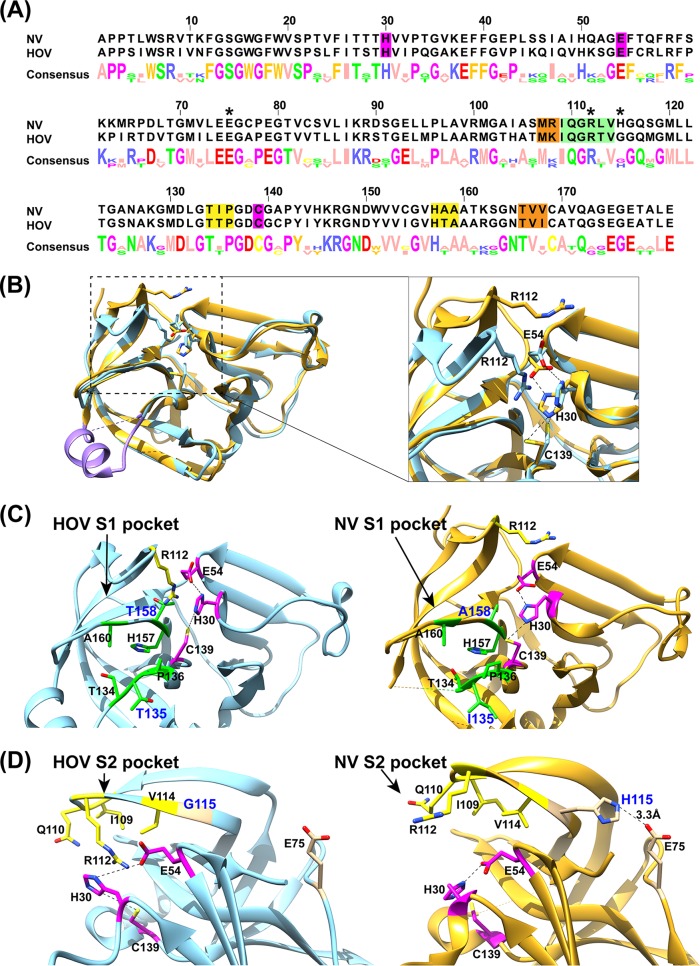
FIG 2.
Sequence and structure comparisons of HOV and NV proteases. (A) Sequence alignment of HOV and NV proteases using Jalview (57). The active site (magenta), and S1 (green), S2 (yellow), and S4 (orange) substrate-binding pockets are highlighted. The residues E75, R112, and H115 are indicated with asterisks. (B) Structural overlay of HOV Pro (light blue) and NV (gold; PDB accession no. 2FYQ) proteases. The residues 122 to 134 that form the floor of the S1 pocket are colored in purple. The inset shows the structural changes of the bII-cII loop between HOV and NV proteases. Active site residues H30, E54, and C139, and the S2 substrate-binding pocket residue R112 are shown as sticks and colored as in panel A. (C and D) Detailed view of the active site and S1 (green) and S2 (yellow) substrate-binding pockets. For clarity, the overlaid structures are shown side by side. R112 in the HOV Pro structure extends into the active site and interacts with H30, while R112 in the NV protease structure is turned away from the active site.