FIG 7.
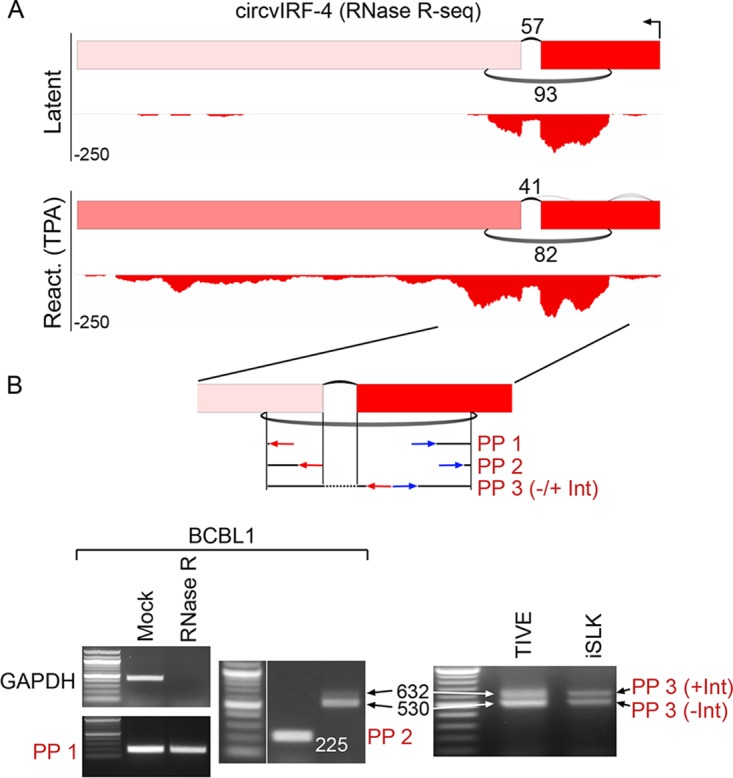
Structure of KSHV circvIRF4 isoforms. (A) Graphical presentation of RNase R-seq splicing and coverage data at the vIRF4 locus in latent and TPA-treated (reactivation) BCBL1 cells. Backsplicing read counts are represented by the number of underarches, forward splicing is displayed by overarches, and exon-level coverage data are indicated based on exon color intensity. Single-nucleotide resolution coverage data are shown below each splicing graph, with amplitudes displayed by negative numbers to represent the leftward orientation of transcription. (B) RNase R resistance and circvIRF4 isoform structure determination. Three different sets of divergent primer pairs were used, with the PP3 primer pair giving rise to two PCR fragments representing a forward-spliced version (530 bp) and an intron-retained version (632 bp) of circvIRF4. Analyses were carried out using naturally KSHV-positive BCBL1 cells and in KSHV-infected TIVE and iSLK cells.
