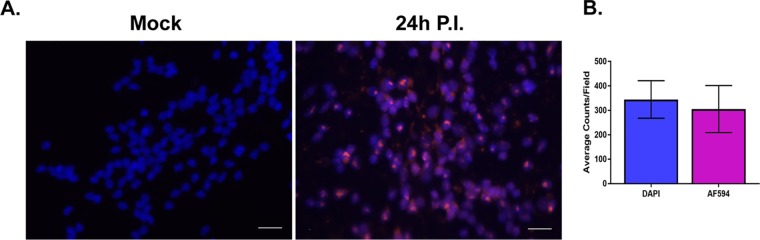
FIG 5.
HSV-1 genomes were characterized by FISH. LUHMES cells were plated and differentiated on coverslips as described in Materials and Methods and infected with 17syn+ at an MOI of 3 in the presence of 50 μM ACV. Coverslips were harvested at 24 h p.i. and neuronal cultures analyzed for the presence of HSV-1 genomes using a biotin-labeled HSV-1 probe followed by amplification with HRP-streptavidin and Alexa Fluor 594 tyramide (AF594). (A) The majority of neuronal nuclei display evidence of harboring HSV-1 genomes (compare mock with 24 h p.i.). Genomes are shown in magenta and DAPI-stained nuclei in blue; scale bar, 20 μm. (B) Total nuclei (DAPI) and replication centers (AF594) were quantified by counting each from 7 fields of cells, and the data are represented as average counts per field. The proportion of neuronal nuclei containing at least one genome-positive signal was 87.4% ± 15.9%.