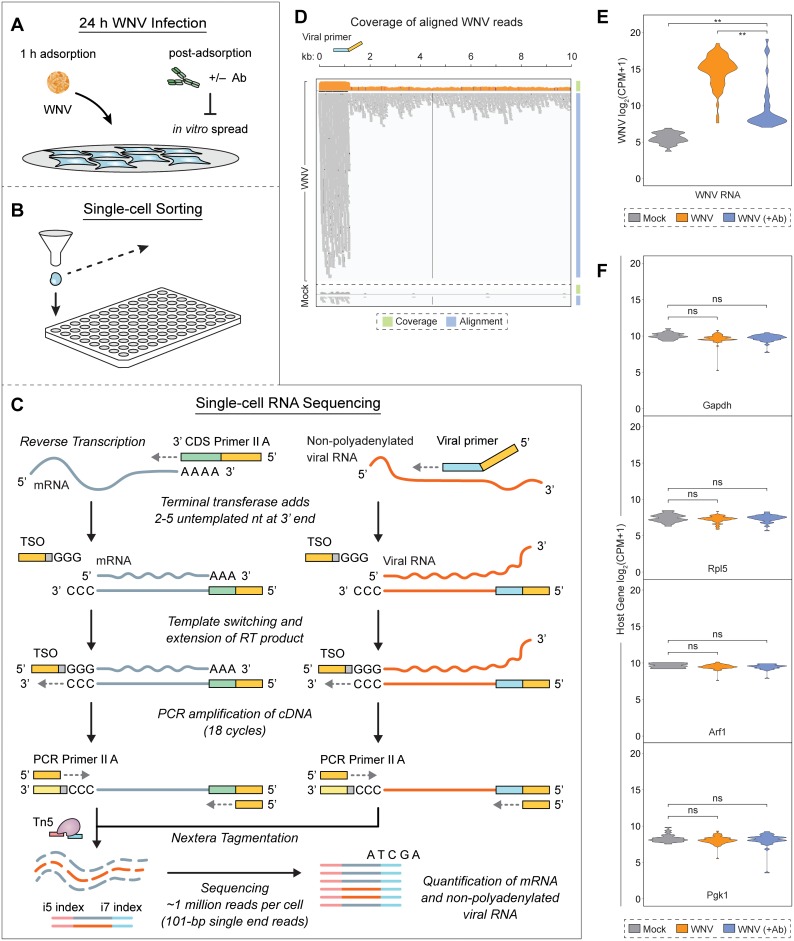
FIG 2.
WNV-inclusive single-cell RNA sequencing. (A) L929 cells were infected with WNV (MOI of 1) and incubated in the presence of the WNV E16 neutralizing Ab (5 μg/ml) to limit in vitro spread. (B) Single cells were sorted into 96-well PCR plates containing 10 μl lysis buffer per well. (C) During reverse transcription (RT), 3′ SMART-Seq CDS Primer II A [30-nucleotide poly(dT) sequence with a 5′ 25-nucleotide ISPCR universal anchor sequence] and a WNV-specific viral primer (21-nucleotide sequence complementary to positive-strand viral RNA with a 5′ 25-nucleotide ISPCR universal anchor sequence) are added to capture host transcripts and viral RNA, respectively. When the reverse transcriptase reaches the 5′ end of both host mRNA and viral RNA, its terminal transferase activity adds two to five untemplated nucleotides that serve as an anchor for the template-switching oligonucleotide (TSO), which allows extension of the RT product with sequence complementary to the universal anchor sequence. PCR Primer II A binds this sequence for concurrent amplification of both host and viral cDNA. In the final library preparation step, transposase 5 (Tn5)-based Nextera tagmentation adds sequencing indices. Illumina sequencing is performed using 101-bp single end reads, thereby quantifying host mRNA and viral RNA from single cells. (B and C) In total, 127 cells were successfully captured and profiled: 25 Mock cells, 68 WNV cells, and 34 WNV (+Ab) cells. (D) Coverage and alignment of WNV reads are shown with reference to the WNV genome and WNV SC primer (viral primer) location, and y axes for coverage are presented in a log10 scale. The cells representing the median value for WNV and Mock conditions are shown. (E) Violin plot showing expression as counts per million transcripts (CPM) in log2 scale for WNV RNA in all three conditions described in panel A. Wilcoxon rank sum test with continuity correction was performed to test significance (**, P < 10−9). (F) Violin plots showing expression as CPM in log2 scale for housekeeping genes. Wilcoxon rank sum test with continuity correction was performed to test significance (ns, not significant; *, P < 10−3; **, P < 10−6).