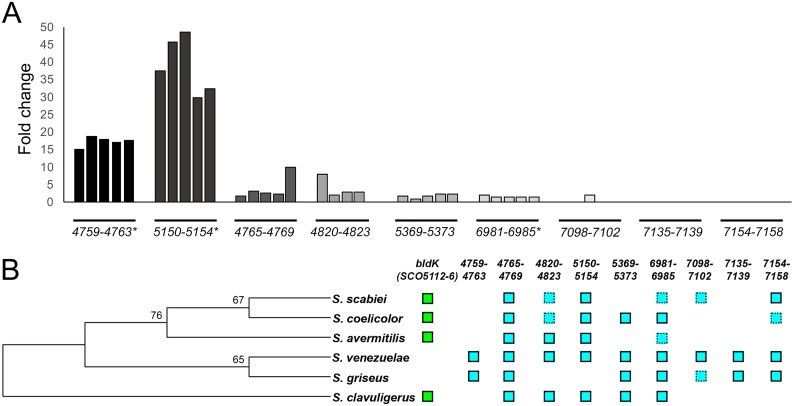
FIG 4.
Upregulation of two gene clusters associated with siderophore transport in explorer cells. (A) Normalized transcript levels for S. venezuelae clusters homologous to the S. coelicolor bldKABCDE locus; values representing normalized transcript levels in S. venezuelae explorer cells were divided by values representing those in nonexploring cells. The associated sven gene numbers are shown below each cluster, and asterisks beside gene numbers indicate that there are genes in the cluster that are significantly differentially expressed in exploring versus nonexploring cultures, based on calculated false-discovery-rate (q) values. (B) Phylogenetic distribution of bldK-like gene clusters in well-studied Streptomyces species. The Streptomyces phylogeny was created using aligned 16S rRNA sequences, and a maximum likelihood tree was built using MEGA, with 100 bootstrap replicates to infer support values of nodes. Green boxes, presence of reciprocally orthologous bldK clusters; turquoise boxes, presence of reciprocally orthologous bldK-like clusters from S. venezuelae; hatched turquoise boxes, presence of orthologues to a subset of genes from each cluster.