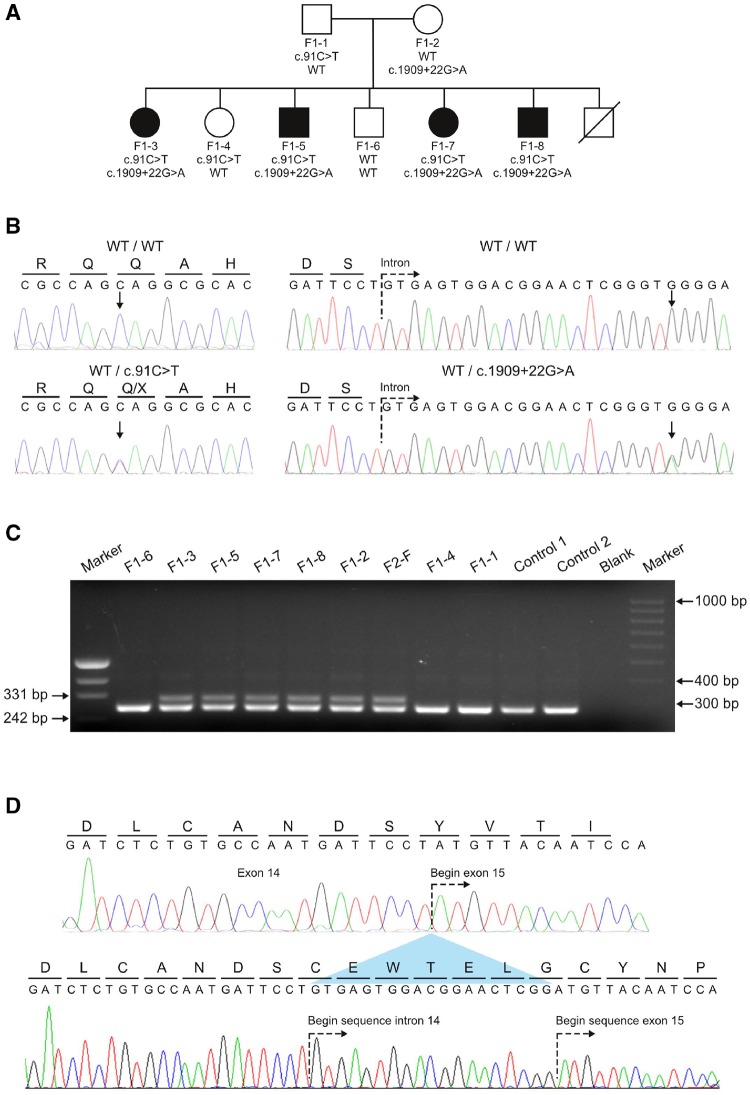
Figure 1.
Genetic screening of a non-consanguineous German family. (A) Pedigree of Family F1 affected by the novel spastic ataxia syndrome. Pedigree symbols: circle, female; square, male; filled, affected; unfilled, unaffected; strike through, deceased. The compound heterozygous mutations found in POLR3A are shown below each symbol. Both mutations segregated with the disease phenotype in the family. (B) Sequencing traces showing the wild-type alleles and the mutant alleles c.91C>T and c.1909+22G>A. Arrows denote the nucleotides undergoing a change for the respective mutations. All affected members in Family F1 are compound heterozygous carriers for both mutations. (C) Agarose gel showing PCR product obtained from the amplification of cDNA template derived from patients, healthy family members, and two controls to show the effect of c.1909+22G>A on the splicing of exon 14. A primer pair flanking exon 14 was used to amplify leucocyte-derived cDNA template from six carriers of the c.1909+22G>A variant and three additional family members of Family F1 that carry the wild-type allele (Patients F1-1, F1-4 and F1-6). An additional aberrant larger fragment (298 bp) in carriers of c.1909+22G>A is not present in the five non-carriers of the splice site mutation. (D) Sequencing analysis of the aberrant amplicon. The upper sequence trace shows the wild-type cDNA sequence obtained for exon 14 and 15. The predicted amino acid translation of this sequence is shown above the nucleotides. The lower sequence trace represents direct sequencing of the aberrant fragment obtained in carriers of the c.1909+22G>A allele. An out-of-frame inclusion of the first 19 nucleotides of intron 14 (caused by the activation of a cryptic splice site) causes a shift of the reading frame.