Figure 4.
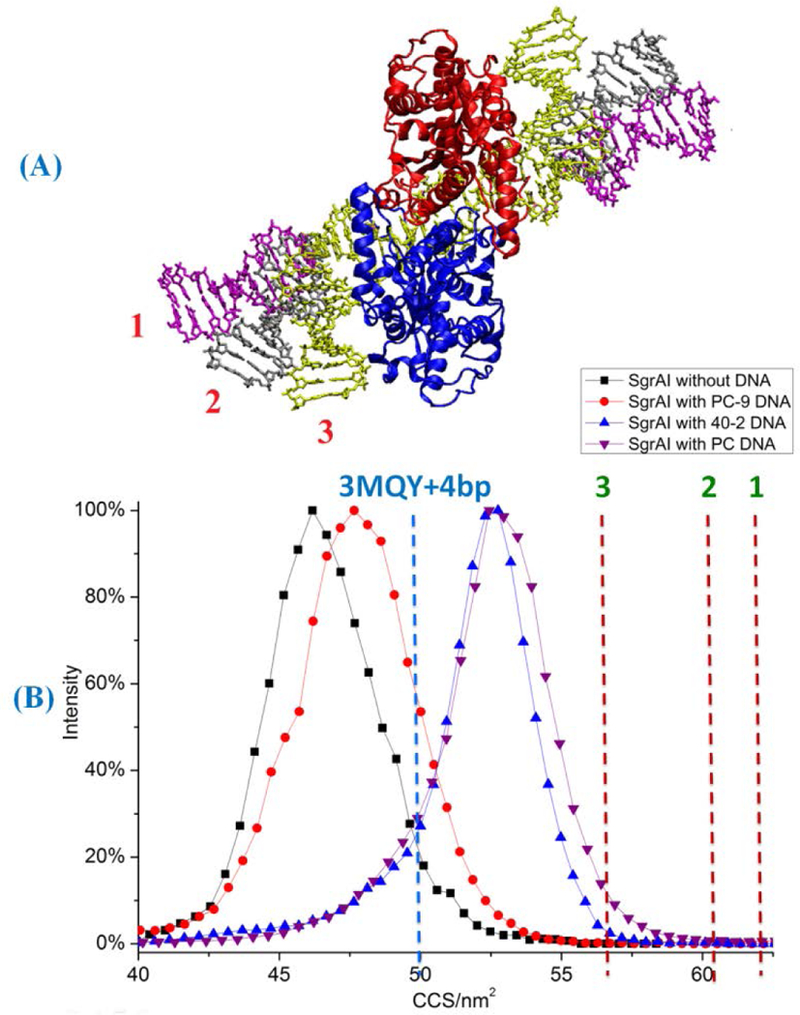
CCSs of SgrAI dimer with different DNA complexes. (A) Three different conformations of flanking DNA (labeled with different colors). 11 bp of B-DNAs were added on both sides of x-ray crystallographic structure 3MQY as flanking DNA. Models with three different flanking DNA conformations were built. In the first model, the flanking DNA is modeled as straight B-form DNA elongated straightly outward into space. In the second model, the flanking DNA bends towards SgrAI by ~30° based on the first model. In the third model, the flanking DNA bends towards SgrAI by ~80°. (B) CCSs of experimental and modeled complexes: black squares, SgrAI dimer without DNA, red circles, SgrAI with PC-9 DNA, blue triangles, SgrAI with 40–2 DNA, purple triangles, SgrAI with PC DNA complex. Dashed lines are calculated CCS of 3MQY with 4 bp of modeled flanking DNA (2 each site of recognition sequence, blue dash line), and calculated CCSs (red dash lines) of models 1–3 (A).
