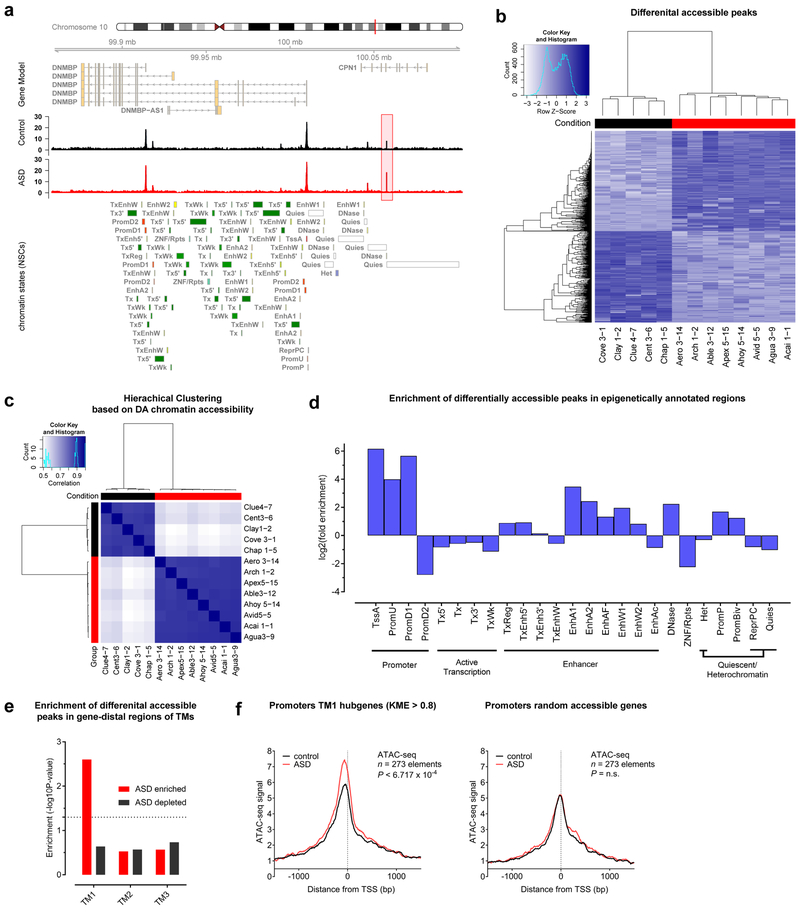
Figure 8: Aberrant gene network dynamics at early neuronal stages are associated with changes in chromatin accessibility at preceding NSC stages.
a, Coverage maps of normalized ATAC-seq signals from ASD and control NSCs showing a differentially accessible (DA) peak (highlighted in red) near the DNMBP gene on chromosome 10. Group-wise sample coverages are displayed and annotated chromatin states are based on the ENCODE 25-state model (see Supplementary Methods). b, Binding affinity heatmap showing normalized accessibilities for DA peaks. c, Correlation heatmap showing hierarchical clustering based on DA peaks (Pearson correlations of peak scores); ASD: n=8 biologically independent patient lines, control: n=5 biologically independent patient lines. d, Log2 fold enrichment of DA peaks in epigenetically annotated regions of the genome (ENCODE 25-state model) shows significant enrichment in promoter and enhancer regions (GAT randomization test53; n=1,593 DA elements; see Supplementary Methods). e, Significance calculations for the enrichment of DA peaks within gene-distal regions (50kbp windows) of TM-associated genes (GAT randomization test53; ASD enriched: n=721 gene-distal DA peaks, ASD depleted: n=755 gene-distal DA peaks; see Supplementary Methods). f, Metagene profiles of normalized ATAC-seq signals at promoters around ±1kb from the transcription start sited (TSS) in NSCs from ASD patients (red) or controls (black). Left: Metagene profiles of promoter regions from TM1 hub genes (kME > 0.8) show higher accessibility in ASD NSCs (P=6.717×10−4, Hotelling’s t test; n=273 elements). Right: Metagene profiles of promoter regions from randomly accessible background genes (P=0.7761, Hotelling’s t test, n.s. – not significant; n=273 elements). Numbers of plotted elements (genes) are similar.