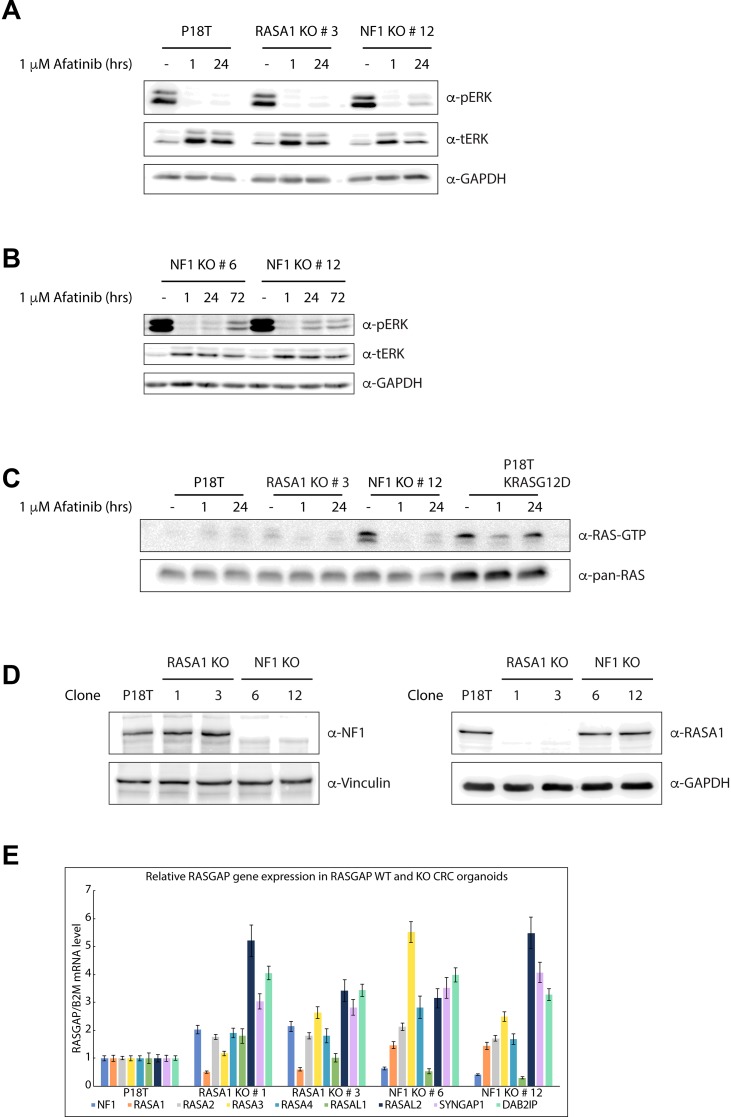
Figure 5. Puromycin selected NF1 knock out CRC organoids show enhanced RAS and ERK activation.
(A) In comparison to P18T and P18T RASA1KO (clone # 3), predominantly P18T NF1KO (clone # 12) organoids show enhanced basal and reactivated ERK phosphorylation levels after 24 hr treatment with CRC medium containing 1 μM afatinib. Representative from n = 3 independent experiments. (B) NF1-deficient organoids (clone # 6 and # 12) show residual ERK phosphorylation after treatment with CRC medium containing 1 μM afatinib with varying kinetics. (C) Loss of NF1 (clone # 12) leads to elevated levels of RAS activity (GTP-loading) at basal conditions compared to P18T and P18T RASA1KO (clone # 3) CRC organoids. The presence of an oncogenic mutation in KRAS (P18T KRASG12D) leads to elevated and sustained high levels of RAS activity (GTP-loading) at basal and in afatinib-treated conditions, respectively. RAS immunoblots from RAS pull-down assays are shown (RAS-GTP), together with a RAS immunoblot from total cell lysates as loading control. HRAS, KRAS, and NRAS isoforms are detected. Representative from n = 2 independent experiments. (D) Immunoblots of P18T, P18T RASA1KO (clone # 1 and 3), P18T NF1KO (clone # 6 and 12) CRC organoids indicate that the loss of RASA1 does not result in elevated protein levels of NF1, and vice versa. Representative from n = 3 independent experiments. (E) The relative expression levels of indicated RASGAPs genes that contain an active GAP domain were analyzed in P18T, P18T RASA1KO (clone # 1 and 3), P18T NF1KO (clone # 6 and 12) CRC organoids using RT-PCR. The relative expression of each RASGAP gene was normalized to the B2M housekeeping gene (representative from n = 3 independent experiments).