Figure 1.
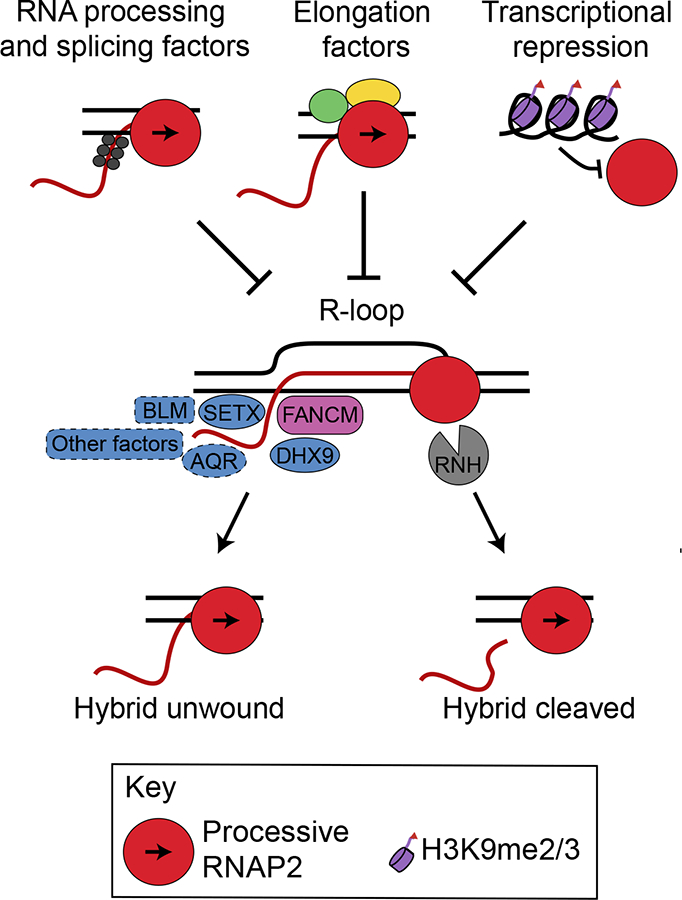
R-loop resolution and suppression mechanisms. R-loops form co-transcriptionally when nascent RNA (red) hybridizes with DNA (black), generating an RNA-DNA hybrid and displaced ssDNA. R-loops are suppressed by processing/splicing factors which coat nascent RNA, elongation factors that ensure processive transcription, or transcriptional repression at certain repeat sequences. Once formed, a variety of helicases (blue) and other factors such as the FANCM translocase, RNase H family nucleases (RNH), or the replisome (not shown) may remove the hybrid, restoring processive transcription. Not all factors implicated in this process have been illustrated for clarity, and many factors have not been examined in biochemical assays to support the proposed activity. Factors outlined in dotted lines show R-loop resolution in cellular assays, but have not been shown to biochemically resolve RNADNA hybrids in vitro.
