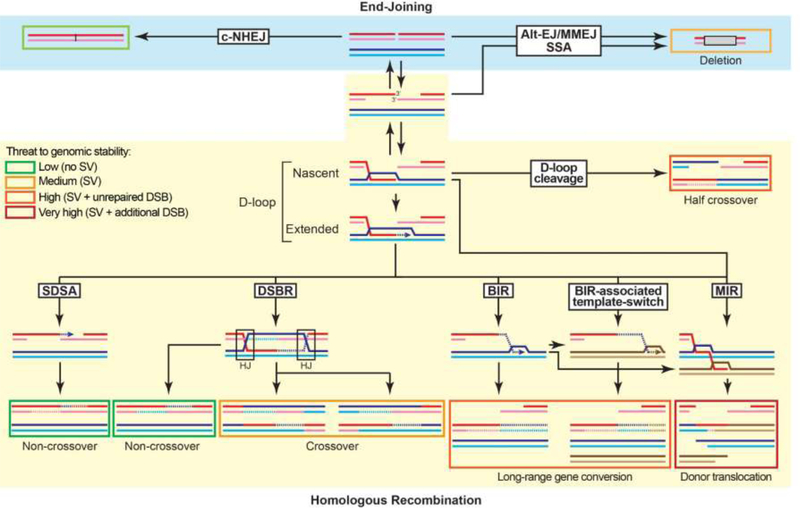
Figure 1: Overview of the DSB repair pathways and their associated risk for genomic stability.
The repair products are boxed. The box color (from green to red) indicates the threat to genomic stability posed by the product of each pathway and sub-pathway. DNA synthesis is indicated by an arrow and newly-synthesized DNA by a dotted line. The long ssDNA associated with BIR has the potential to undergo MIR. More detailed mechanisms for D-loop cleavage and MIR are provided in Figures 2A and 2C, respectively. The reversibility of DSB resection provided by the Shieldin complex with fill-in by the DNA polymerase α-primase complex provides an unanticipated degree of flexibility in the choice of DSB repair between HR and EJ mechanisms [130]. Hence, EJ pathways are available not only on unresected or minimally resected DSB, they can also be engaged after extensive resection and subsequent fill-in.