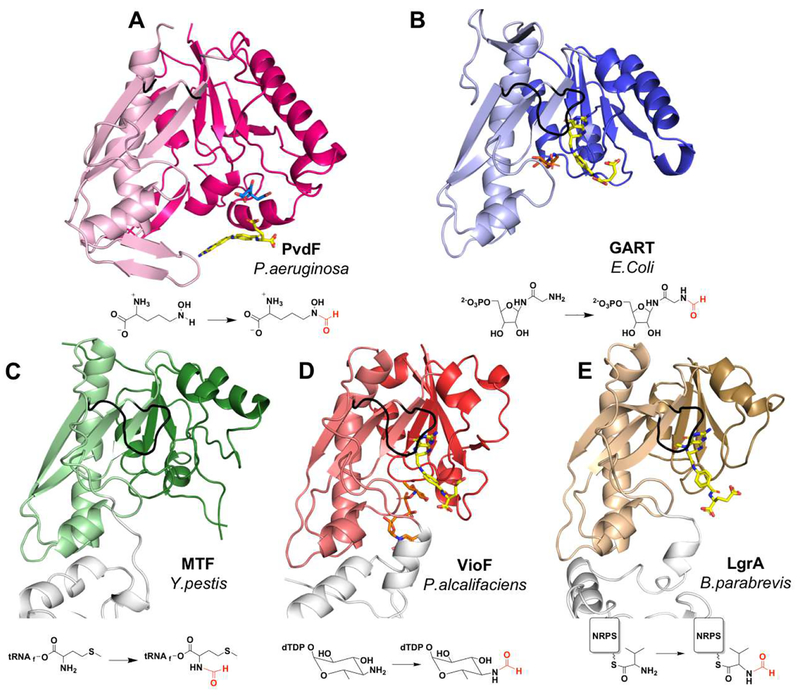
Figure 4.
Comparison of PvdF with structural and functional homologues. (A) PvdF (B) GART from E.Coli (PDB:1CDE; sequence identity when compared to PvdF: 27%, calculated in LALIGN); (C) methionyl t-RNA transformylase from Y.pestis (PDB:3R8X; seq id: 26%); (D) VioF, sugar N-transformylase from P.alcalifaciens (PDB:4YFY; seq id: 28%); and (E) the NRPS formyltransferase domain of LgrA from B.parabrevis (PDB:5ES7; seq id: 25%). In each case the darker shade is the N-terminal and lighter shade is the C-terminal domain. In the bottom row the transformylase is part of a large multi-functional enzyme and domains without transformylase activity are white. The active site loop is shown in black. Folate analogues are shown in yellow with respective substrates in orange. The citrate molecule from crystallization in PvdF is shown in cyan. The reaction catalyzed by each enzyme is represented under the structure.